Detection of Alternative Splicing During Epithelial-Mesenchymal Transition
Summary
Alternative splicing regulation has been shown to contribute to the epithelial-mesenchymal transition (EMT), an essential cellular program in various physiological and pathological processes. Here we describe a method utilizing an inducible EMT model for the detection of alternative splicing during EMT.
Abstract
Alternative splicing plays a critical role in the epithelial-mesenchymal transition (EMT), an essential cellular program that occurs in various physiological and pathological processes. Here we describe a strategy to detect alternative splicing during EMT using an inducible EMT model by expressing the transcription repressor Twist. EMT is monitored by changes in cell morphology, loss of E-cadherin localization at cell-cell junctions, and the switched expression of EMT markers, such as loss of epithelial markers E-cadherin and γ-catenin and gain of mesenchymal markers N-cadherin and vimentin. Using isoform-specific primer sets, the alternative splicing of interested mRNAs are analyzed by quantitative RT-PCR. The production of corresponding protein isoforms is validated by immunoblotting assays. The method of detecting splice isoforms described here is also suitable for the study of alternative splicing in other biological processes.
Introduction
The epithelial-mesenchymal transition (EMT) is a developmental program that drives organ morphogenesis and tissue remodeling during embryogenesis. When abnormally activated, EMT promotes tumor metastasis and organ fibrosis1,2. Compelling studies have described the importance of transcriptional regulation during the process of EMT, defined by several transcription factors, such as Twist, Snail, and ZEB, which repress the expression of the adherens junction protein E-cadherin, resulting in loss of a cobble-stone like epithelial morphology and gain of a spindle-shaped mesenchymal phenotype3-8. Recent studies through genome-wide analysis of RNAs revealed that there exists a group of genes whose splicing patterns are associated with either epithelial or mesenchymal phenotypes9,10. Work from our lab functionally connected alternative splicing and EMT. By studying the cell surface adhesion molecule CD44, we demonstrated that CD44 alternative splicing is tightly regulated during EMT, and more importantly, that CD44 splice isoform switching causally contributes to EMT11.
Alternative splicing represents a widespread and conserved model of gene regulation, as up to 95% of human multi-exon genes are alternatively spliced12-14. By generating multiple protein products from a single gene, alternative splicing constitutes an essential mechanism for protein diversity, adding another layer of complexity to the human genome. As such, dysregulation of alternative splicing could potentially lead to profound biological effects causing human diseases. Indeed, aberrant alternative splicing in diseases has been documented for over a decade15-25, including recent findings that mutations in genes encoding the spliceosome machinery are commonly found in myelodysplastic syndromes26-28. Therefore, developing reliable methods for the detection of alternatively spliced isoforms is of great importance in the study of diverse biological processes including EMT.
Here we provide a protocol to detect changes in alternative splicing using an inducible EMT model. The methods for designing PCR primers and detecting splice isoforms are suitable not only for the study of alternative splicing during EMT, but also for the study of alternative splicing in other biological processes. Investigating alternative splicing during EMT is imperative in order to better understand the mechanisms of EMT and tumor metastasis, thus facilitating the development of effective strategies to treat cancer metastasis.
Protocol
1. Cell Culture of EMT Induction
Note: EMT can be induced by treatment of TGFβ or ectopic expression of transcription factors Twist, Snail, or Zeb1/2 in epithelial cells. Described in this protocol is an inducible EMT system via expression of the Twist-ER fusion protein in immortalized human mammary epithelial cells (HMLE/Twist-ER, a gift of Dr. J Yang, UCSD)9,11,29. Upon 4-hydroxytamoxifen (TAM) treatment, the fusion protein Twist-ER translocates to the nucleus to drive transcription and results in a full EMT transition in 12-14 days9,11,29. Additional EMT inducible systems, such as HMLE/Snail-ER, HMLE/TGFβ, and MCF10A/TGFβ, can be found in our previous publications11,30.
- Maintain HMLE/Twist-ER cells in serum-free mammary epithelial cell growth medium (MEGM) in a 37 °C incubator with 5% CO2. Passage the cells every 2-3 days when they reach 80% confluence. Incubate cells with 0.15% trypsin at 37 °C for 5-10 min, and then inactivate trypsin with 5% calf serum/DMEM. Spin down cells and resuspend them in MEGM. Plate cells in a new tissue culture dish at a ratio of 1 to 4.
- Dissolve TAM in ethanol to make a 200 μM solution. Protect TAM from light and store at -20 °C. Before use, freshly add TAM into MEGM media at a 1:10,000 dilution to make a final concentration of 20 nM TAM.
- For induction of EMT, plate 1.6 x 106 HMLE/Twist-ER cells into a 10 cm tissue culture dish in duplicate. Culture cells with 20 nM TAM-containing MEGM media.
- Plate 1.6 x 106 HMLE/Twist-ER cells into a 10 cm dish in duplicate and treat cells with 0.01% ethanol as a non-TAM-treated control.
- Two days after incubation, take pictures under a light microscope at 10X magnification to record morphological changes of cells.
- Aspirate the medium from one dish of control or TAM-treated cells. Wash cells with cold PBS, and harvest cells by scrapping half of the plate in RNA lysis buffer for RNA isolation and the other half in RIPA buffer for protein analysis.
- Passage cells from the control or TAM-treated dishes by re-plating 1.6 x 106 cells in a 10 cm dish in duplicate. Continuously treat the cells with 20 nM TAM or vehicle.
- Repeat steps 5 to 7 once every other day until day 14, at which time, TAM-treated Twist-ER cells show a spindle-shaped mesenchymal morphology, whereas vehicle-treated control cells maintain the cobblestone-like epithelial morphology.
2. Characterization of EMT
Note: A completion of EMT is indicated by: (1) Cell morphological changes from a cobblestone-like epithelial morphology to a spindle-shaped fibroblastic-like appearance; (2) Loss of E-cadherin localization at cell-cell junctions; and (3) The switched expression of EMT markers, defined by the loss of epithelial markers and gain of mesenchymal markers.
Cell morphological changes are captured by light microscope at 10X magnification during the time course of EMT induction, described in Section 1. Immuno-fluorescence detection of E-cadherin at cell-junctions is described in this section. The expression of EMT markers is detected by immunoblotting. Common epithelial markers are E-cadherin, γ-catenin, and occludin, and mesenchymal markers include fibronectin, N-cadherin and vimentin. General procedures of immunoblotting are described in Section 4.
- At day 12 of TAM treatment, seed 1 x 105 cells on a 12 mm glass circular coverslip placed in a well of a 24-well plate.
- 48 hr after seeding, aspirate medium and wash cells with pre-warmed PBS.
- Fix cells with pre-warmed 4% paraformaldehyde for 15 min.
- Wash cells three times with PBS.
- Incubate cells with 0.2% Triton X-100 in PBS for 15 min at RT.
- Wash cells three times with PBS.
- Incubate cells in 5% BSA/PBS for 1 hr at RT to block non-specific binding.
- Incubate cells with the primary antibody E-cadherin at a 1:50 dilution in 1% BSA/PBS in a humidified chamber O/N at 4 °C. NOTE: The range for primary antibody dilution is normally between 1:50 and 1:200, and the optimal dilution for each antibody should be determined experimentally.
- Decant the primary antibody and wash cells three times with PBS.
- Incubate cells with fluorescent-labeled secondary antibody in a 1:500 dilution for 1 hr at RT. From this step on, protect cells from light.
- Decant the secondary antibody and wash three times with PBS.
- Stain cells with 0.1-1 g/ml DAPI or Hoechst for 10 min.
- Rinse cells with PBS three times.
- Mount coverslips with a drop of mounting medium, and seal coverslips with nail polish.
- Observe cells and take images under a 63X fluorescent microscope.
Staining with additional antibodies can be performed to confirm the EMT phenotype. For example, N-cadherin and α-smooth muscle actin can be stained for a mesenchymal phenotype29,31,32. Reorganization of cytoskeletal structure can be monitored by staining cells with phalloidin for F-actin11. In addition, assays for cell motility and cell-death resistance can be performed to examine the properties of EMT11.
3. Detection of Splice Isoforms Using qRT-PCR Assays
- Primer Design
NOTE: Primer pairs should be carefully designed to amplify distinct splice isoforms. Exon skipping, the most commonly observed alternative splicing event, is shown here as an example to illustrate the strategy of primer design. Figure 1A depicts a four-exon pre-mRNA, with the third exon as a variable exon. Inclusion of exon 3 results in the production of a longer splice isoform, whereas skipping of exon 3 generates a shorter splice isoform. Primers need to be designed to distinguish the exon-3-included isoform and exon-3-skipped isoform, and to detect the total transcript of this mRNA.- For detection of exon inclusion, carefully design a forward primer inside the variable exon 3, and a reverse primer inside the nearby constitutive exon 4, allowing for the specific amplification of the variable exon-included isoform. Make sure not to design both forward and reverse primers within the same variable exon in order to avoid PCR products amplified from genomic DNA or pre-mRNA.
- To specifically amplify exon skipping events, design one of the primers spanning the junction of the constitutive exon 2 and exon 4 that would otherwise be destroyed when variable exon 3 is included. Design the other primer inside the constitutive exon 4. As such, PCR products are generated only when the variable exon 3 is excluded and the constitutive exon 2 and exon 4 are subsequently joined together.
- To detect the total transcripts of the gene, design the primers within two constitutive exons 1 and 2. Thus, PCR products are amplified from all isoforms.
- Make sure the melting temperature (Tm) for all primers is approximately 55 °C, the length of each primer is approximately 18-22 nucleotides, and the size of PCR products is between 80 and 150 base pairs.
- RNA extraction: Extract RNA using a total RNA isolation kit (see Table of Materials).
- Collect cells in 350 μl RNA lysis buffer.
- Add 350 μl of 70% ethanol to the lysate and mix thoroughly.
- Transfer the sample to the RNA purification column.
- Centrifuge at 10,000 x g for 1 min and discard flow-through.
- Wash the column once with 500 μl of RNA wash buffer I and twice with RNA wash buffer II.
- Remove residual RNA wash buffer II from the column by centrifugation at maximum speed for 2 min.
- Transfer the column into a new 1.5 ml tube, add 50 μl nuclease-free water at the center of the column matrix and elute RNA by centrifugation at the maximum speed.
- Synthesis of First-strand cDNA
- Determine the concentration and quality of RNA by UV absorption measurement. NOTE: Good quality RNA should have a 260 nm / 280 nm absorbance ratio of approximately 2.0.
- Set up reverse transcriptase (RT) reactions that contain 250 ng total RNA, 0.05 μg random hexamer primers, 50 pmol MgCl2, 10 pmol dNTP, 2 μl 5× GoScript buffer, and 1 μl RT enzyme in a final volume of 20 μl.
- Perform RT reactions at 42 °C for 1 hr, followed by incubation at 70 °C for 5 min to inactivate the RT enzyme.
- Real-time PCR
- Set up 20 μl reactions that consist of 10 μl SYBR green master mix, 0.1-1.0 μl cDNA template, and 5 pmol forward and reverse primers.
- Run real-time PCR with 40 cycles of the following steps: 95 °C denaturation for 10 sec, 58 °C annealing for 10 sec, and 60 °C extension for 30 sec. Perform PCR reactions in triplicate.
- Check dissociation curves and make sure that a single peak is observed for each primer set.
- Obtain the quantification cycle (Cq) values by calculating the second derivative values of amplification curves.
- Calculate the relative quantity (RQ) of target mRNAs in induced samples as compared to the uninduced sample using the ‘delta delta Cq (ΔΔCq)’ method as shown by the following formula. Use housekeeping genes, such as GAPDH or TBP, as a reference.
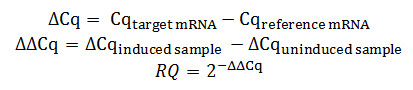
To more accurately calculate relative values of splice isoforms, the use of multiple reference genes should be considered. The results can be analyzed using a modified absolute value quantification method described by Pfaffl et al33,34.
4. Examination of Splice Isoforms at the Protein Level
- Collect cells in 100 μl ice-cold RIPA buffer.
- Set lysates on ice for approximately 10 min.
- Centrifuge at 14,000 x g for 10 min at 4 °C, and collect the supernatant.
- Measure protein concentration by Bradford assays.
- Dilute protein samples in SDS loading buffer and load 20-40 μg of proteins into 10% SDS-PAGE gels.
- Run SDS-PAGE gels at 20 mA for 1.5 hr.
- Transfer proteins to PVDF membranes in a wet transfer system at 4 °C for 3 hr at 85 V. Adjust the transfer time according to the molecular weight of target proteins.
- Block membrane in 5% non-fat milk for 30 min to 1 hr at RT.
- Incubate membrane with a primary antibody at 4 °C O/N. The range for primary antibody dilution is normally between 1:200 and 1:5,000, determine the optimal dilution for each antibody experimentally. The dilution of antibodies used in this protocol can be found in “Table of specific reagents and equipment”.
- To detect splice isoforms, use specific antibodies that recognize a particular isoform. Alternatively, use an antibody that recognizes the epitope in the constitutive exon coding region and detect splice isoforms simultaneously based on their different protein sizes.
- At the same time, monitor EMT by immunoblotting for EMT markers. Use E-cadherin, γ-catenin, and occludin as epithelial markers, and fibronectin, N-cadherin and vimentin as mesenchymal markers.
- Wash membrane three times with TBS-T (50 mM Tris, 150 mM NaCl, 0.05% Tween 20, pH 7.4) at a 5-min interval.
- Incubate membrane with an HRP-conjugated secondary antibody in a 1:10,000 dilution for 1 hr at RT.
- Wash membrane three times with TBS-T at a 5-min interval.
- Visualize protein with a chemiluminescence detection system and expose the membrane to an autoradiography film.
Representative Results
The procedures described above provide a robust method to detect alternative splicing during EMT. Representative results of CD44 splice isoform switching during Twist-induced EMT are given below as an example.
Twist-induced EMT in HMLE/Twist-ER cells was characterized by a transition from a cobble-stone like epithelial phenotype to an elongated fibroblastic phenotype (Figure 2A), the absence of epithelial markers E-cadherin, γ-catenin and occludin and the upregulation of mesenchymal markers fibronectin, N-cadherin, and vimentin (Figure 2B). Furthermore, EMT was assessed by the loss of E-cadherin localization at cell-cell junctions (Figure 2C).
The switched expression of CD44 splice isoforms during EMT was examined by qRT-PCR and immunoblotting. The human CD44 gene is comprised of nine variable exons. Alternative splicing of CD44 allows for cells to produce CD44 variants (CD44v) that include at least one variable exon, and CD44 standard (CD44s) that is devoid of all variable exons. Figure 3A depicts the strategy of primer design for the detection of various CD44 splice isoforms. For the detection of the exon-skipped product CD44s, the forward primer is located in the constitutive exon 5, while the reverse primer spans across the junction between constitutive exons 5 and 6. For the detection of CD44v splice isoforms containing the v5 and v6 variable exons, the forward and reverse primers are designed within the v5 and v6, respectively. Primers for detection of other CD44 variable exon-containing isoforms can be designed using the same strategy. Additionally, total CD44 transcript is detected using forward and reverse primers located in the constitutive exon 2 and exon 3, respectively (Figure 3A). As shown in Figure 3B, qRT-PCR analyses utilizing these primer sets indicated a significant decrease in CD44v mRNA and increase in CD44s mRNA after 14 days of TAM treatment in the Twist-ER-expressing HMLE cells. By contrast, the total transcript of CD44 remained unchanged during EMT (Figure 3C). Consistent with the results of qRT-PCR, the protein level of CD44v declined remarkably, whereas expression of the CD44s protein was greatly upregulated (Figure 3D). Therefore, the major isoform of CD44 was shifted from CD44v to CD44s during the EMT process.
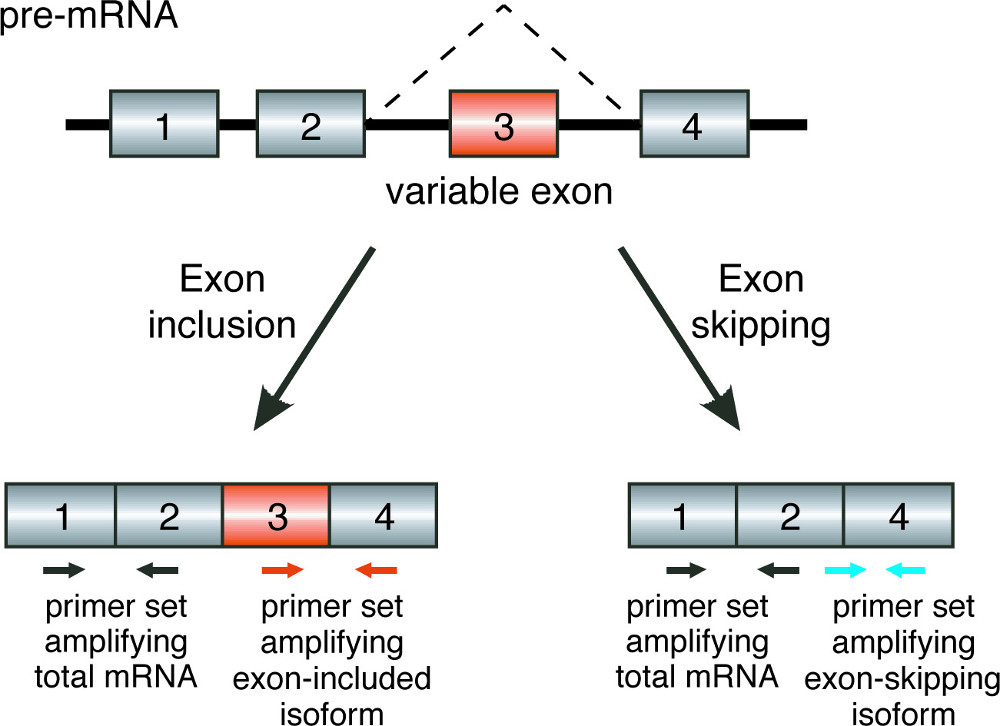
Figure 1. Primer design. Schematic diagrams illustrating the location of primers for detecting splicing isoforms in an exon-skipping model. The gray boxes represent constitutive exons, the orange boxes represent variable exons, and the thin lines connecting boxes denote introns. The primer locations are indicated by arrows: black arrows indicate primer sets for detecting total mRNA, orange arrows indicate primer sets for amplifying exon-included isoforms, and blue arrows indicate primer sets for detecting exon-skipping isoforms.

Figure 2. Induction of EMT. (A) Phase contrast images (10X) illustrating morphological changes in HMLE/Twist-ER cells before (untreated) and after 14 days of TAM treatment. (B) Immunoblot analysis of EMT markers in HMLE/Twist-ER cells before (untreated) and after 14 days of TAM treatment. Upon TAM treatment, epithelial markers E-cadherin, γ-catenin, and occludin were downregulated, and mesenchymal markers fibronectin, N-cadherin, and vimentin were upregulated. (C) Immune fluorescence images (63X) indicating the loss of E-cadherin localization at cell-cell junctions after 14 days of TAM treatment. Green staining indicates E-cadherin, and DAPI staining (blue) indicates nuclei. Scale bar = 10 μm. Please click here to view a larger version of this figure.
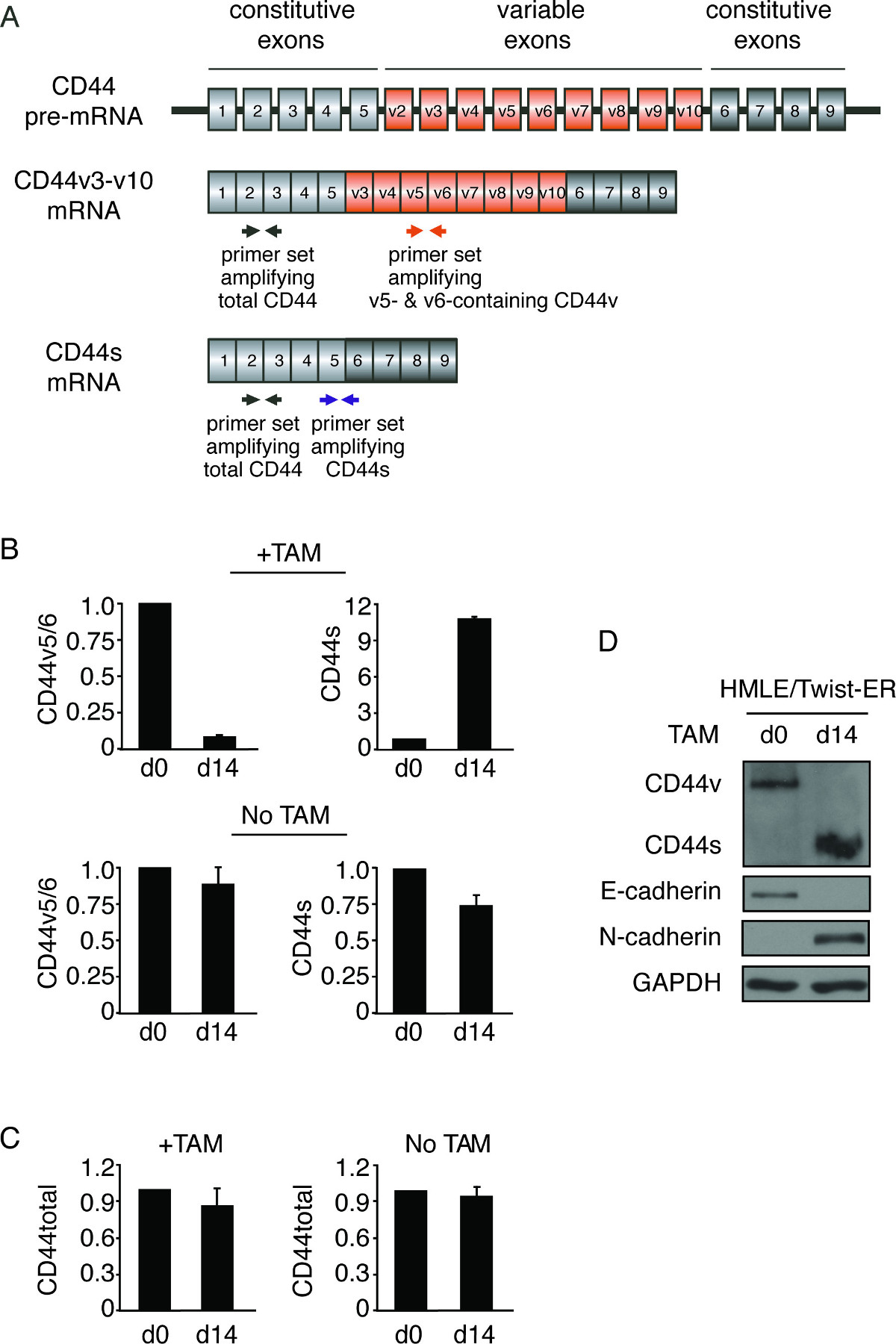
Figure 3. Switch of CD44 splice isoforms during EMT. (A) Primer design for the detection of CD44 splice isoforms. The gray boxes represent constitutive exons, the orange boxes represent variable exons, and the thin lines connecting boxes denote introns. The primer locations are indicated by arrows: black arrows indicate primer sets for detecting CD44 total mRNA, orange arrows indicate primer sets for amplifying CD44v5-6, and blue arrows indicate primer sets for detecting CD44s. (B, C) qRT-PCR analysis of the levels of CD44 isoforms during EMT using primers that specifically detect CD44v containing variable exons v5 and v6 (v5/6) and CD44s (B), and CD44 total mRNA (C). Relative expression levels of mRNA in Day 14 cells were normalized to corresponding day 0 cells in TAM treated and non-treated groups. Error bars indicate SEM; n = 3. (D) Immunoblot analysis of CD44 isoforms during TAM-induced EMT in HMLE/Twist-ER cells. Levels of E-cadherin and N-cadherin were monitored to identify EMT status.
Discussion
The procedure described here enables the detection of alternative splicing in an inducible EMT model. As such, dynamic alterations of splice isoform expression can be captured throughout the time course of EMT. This method has advantages over the use of different epithelial- or mesenchymal-representing cell lines for comparison of alternative splicing because distinct genetic backgrounds from un-related cell lines could unduly influence alternative splicing. However, the successful induction of EMT must be carefully confirmed using various EMT markers before any conclusions on changes in alternative splicing can be drawn. Furthermore, in addition to the Twist-ER-inducible EMT system described here, other EMT systems such as those induced by Snail-ER or TGFβ are recommended for validation of experimental findings11,30.
The switching of splice isoforms during EMT can be detected in both RNA and protein levels. Splice isoform-specific antibodies are preferred for protein analysis. When isoform-specific antibodies are not available, protein levels of splice isoforms can be determined based on their molecular weight on SDS-PAGE by immunoblotting. The RNA level of each isoform can be quantified by isoform-specific primers. Using qRT-PCR assays, the fold change of each isoform can be sensitively and precisely determined. In particular, when experimental materials are limited, such as analyzing the expression of a particular splice isoform in clinical specimens, qRT-PCR analysis provides a quantitative measure that would compensate for the lack of isoform-specific antibodies for immunohistochemistry.
The method mentioned here is suitable for the detection of known splice isoform changes during EMT. Additionally, this method can be expanded for the study of genome-wide alternative splicing and for the identification of novel splice isoform switching during EMT. To accomplish this, use of a large-scale technique, such as RNA deep-Sequencing or splicing sensitive microarray platforms, could be performed using RNAs isolated from the inducible EMT model described above. Nevertheless, qRT-PCR validation of isoform changes is required following any large-scale analysis.
In conclusion, alternative splicing is dynamically regulated during EMT, a process that is critical for embryonic development and tumor metastasis. Considering the high frequency of alternative splicing in the human genome, it is likely that the regulation of alternative splicing plays an important role in many other biological and pathological processes including cell differentiation, tissue development, and programmed cell death35-41. The protocol described herein for the detection of splice isoforms will be applicable to these other systems as well.
Additional experimental modalities, such as splicing reporter minigene assays, can be used to further investigate the regulatory mechanisms of cis-acting elements and trans-acting factors that control alternative splicing30,42,43. Furthermore, the functional role of a particular splice isoform during EMT can be investigated by silencing or ectopically expressing the specific isoform and monitoring EMT induction in the described EMT-inducible system11.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge Wensheng Liu for invaluable help with cell imaging. This work was supported by grants from the US National Institutes of Health (R01 CA182467), American Cancer Society (RSG-09-252-01-RMC), Lynn Sage Foundation, and A Sister’s Hope Foundation.
Materials
| Name of the reagent | Company | Catalogue number | Comments (optional) |
| 4-hydroxytamoxifen | Sigma | H7904 | Make stock solution by dissolving in ethanol to 200μM, and keep at -20℃ protected from light. |
| E.Z.N.A. Total RNA Isolation kit | Omega Bio-Tek | R6731 | Total RNA isolation kit |
| GoScript Reverse Transcription System | Promega | A5001 | Reagent for qRT-PCR assay |
| GoTaq qPCR Master Mix | Promega | A6002 | Reagent for qRT-PCR assay |
| LightCycler 480 Real-Time PCR System | Roche | Equipment for qRT-PCR assay | |
| CD44 antibody | R&D Systems | BBA10 | 1:1000 dilution |
| E-cadherin antibody | Cell Signaling Technology | 4065 | 1:2500 dilution for immunoblotting; 1:50 dilution for immunofluorescence |
| γ-catenin antibody | Cell Signaling Technology | 2309 | 1:1000 dilution |
| occludin antibody | Santa Cruz Biotechnology Inc. | sc-5562 | 1:500 dilution |
| fibronectin antibody | BD Transduction Laboratories | 610077 | 1:5000 dilution |
| N-cadherin antibody | BD Transduction Laboratories | 610920 | 1:2000 dilution |
| vimentin antibody | NeoMarkers | MS-129-p1 | 1:500 dilution |
| GAPDH antibody | Millipore Corporation | MAB374 | 1:10000 dilution |
| Amasham ECL Western blotting detection reagent | GE Health Life Science | RPN2209 | Chemiluminescence system |
References
- Thiery, J. P., Sleeman, J. P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 7, 131-142 (2006).
- Yang, J., Weinberg, R. A. Epithelial-mesenchymal transition at the crossroads of development and tumor metastasis. Dev Cell. 14, 818-829 (2008).
- Yang, J., et al. a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 117, 927-939 (2004).
- Batlle, E., et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2, 84-89 (2000).
- Cano, A., et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2, 76-83 (2000).
- Comijn, J., et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 7, 1267-1278 (2001).
- Bolos, V., et al. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 116, 499-511 (2003).
- Eger, A., et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 24, 2375-2385 (2005).
- Shapiro, I. M., et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 7, e1002218 (2011).
- Warzecha, C. C., et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 29, 3286-3300 (2010).
- Brown, R. L., et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 121, 1064-1074 (2011).
- Wang, E. T., et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 456, 470-476 (2008).
- Pan, Q., Shai, O., Lee, L. J., Frey, B. J., Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 40, 1413-1415 (2008).
- Liu, S., Cheng, C. Alternative RNA splicing and cancer. Wiley Interdiscip Rev RNA. 4, 547-566 (2013).
- Shtivelman, E., Lifshitz, B., Gale, R. P., Roe, B. A., Canaani, E. Alternative splicing of RNAs transcribed from the human abl gene and from the bcr-abl fused gene. Cell. 47, 277-284 (1986).
- Krangel, M. S. Secretion of HLA-A and -B antigens via an alternative RNA splicing pathway. J Exp Med. 163, 1173-1190 (1986).
- Brickell, P. M., Latchman, D. S., Murphy, D., Willison, K., Rigby, P. W. Activation of a Qa/Tla class I major histocompatibility antigen gene is a general feature of oncogenesis in the mouse. Nature. 306, 756-760 (1983).
- Barbaux, S., et al. Donor splice-site mutations in WT1 are responsible for Frasier syndrome. Nat Genet. 17, 467-470 (1997).
- Charlet, B. N., et al. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell. 10, 45-53 (2002).
- Hutton, M., et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 393, 702-705 (1998).
- Kohsaka, T., et al. Exon 9 mutations in the WT1 gene, without influencing KTS splice isoforms, are also responsible for Frasier syndrome. Hum Mutat. 14, 466-470 (1999).
- Philips, A. V., Timchenko, L. T., Cooper, T. A. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 280, 737-741 (1998).
- Savkur, R. S., Philips, A. V., Cooper, T. A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 29, 40-47 (2001).
- Spillantini, M. G., et al. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc Natl Acad Sci USA. 95, 7737-7741 (1998).
- Wang, J., et al. Myotonic dystrophy evidence for a possible dominant-negative RNA mutation. Hum Mol Genet. 4, 599-606 (1995).
- Graubert, T. A., et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 44, 53-57 (2012).
- Papaemmanuil, E., et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 365, 1384-1395 (2011).
- Yoshida, K., et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 478, 64-69 (2011).
- Mani, S. A., et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 133, 704-715 (2008).
- Reinke, L. M., Xu, Y., Cheng, C. Snail represses the splicing regulator epithelial splicing regulatory protein 1 to promote epithelial-mesenchymal transition. J Biol Chem. 287, 36435-36442 (2012).
- Serini, G., Gabbiani, G. Mechanisms of myofibroblast activity and phenotypic modulation. Exp Cell Res. 250, 273-283 (1999).
- Zavadil, J., Bottinger, E. P. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 24, 5764-5774 (2005).
- Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
- Hellemans, J., Mortier, G., De Paepe, A., Speleman, F., Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8, R19 (2007).
- Boutz, P. L., et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 21, 1636-1652 (2007).
- Salomonis, N., et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci USA. 107, 10514-10519 (2010).
- Calarco, J. A., et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 138, 898-910 (2009).
- Grabowski, P. Alternative splicing takes shape during neuronal development. Curr Opin Genet Dev. 21, 388-394 (2011).
- Seol, D. W., Billiar, T. R. A caspase-9 variant missing the catalytic site is an endogenous inhibitor of apoptosis. J Biol Chem. 274, 2072-2076 (1999).
- Srinivasula, S. M., et al. Identification of an endogenous dominant-negative short isoform of caspase-9 that can regulate apoptosis. Cancer Res. 59, 999-1002 (1999).
- Akgul, C., Moulding, D. A., Edwards, S. W. Alternative splicing of Bcl-2-related genes functional consequences and potential therapeutic applications. Cell Mol Life Sci. 61, 2189-2199 (2004).
- Cooper, T. A. Use of minigene systems to dissect alternative splicing elements. Methods. 37, 331-340 (2005).
- Cheng, C., Sharp, P. A. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol Cell Biol. 26, 362-370 (2006).

