Deferred Growth Inhibition Assay to Quantify the Effect of Bacteria-derived Antimicrobials on Competition
Summary
The deferred growth inhibition assay can be used to assess the competition effect by one bacterial isolate on another. Inhibition is quantified by measuring the zone of clearing around the inhibitor-producing isolate, or qualitatively assessed by determining the visible extent of inhibition.
Abstract
Competitive exclusion can occur in microbial communities when, for example, an inhibitor-producing strain outcompetes its competitor for an essential nutrient or produces antimicrobial compounds that its competitor is not resistant to. Here we describe a deferred growth inhibition assay, a method for assessing the ability of one bacterium to inhibit the growth of another through the production of antimicrobial compounds or through competition for nutrients. This technique has been used to investigate the correlation of nasal isolates with the exclusion of particular species from a community. This technique can also be used to screen for lantibiotic producers or potentially novel antimicrobials. The assay is performed by first culturing the test inhibitor-producing strain overnight on an agar plate, then spraying over the test competitor strain and incubating again. After incubation, the extent of inhibition can be measured quantitatively, through the size of the zone of clearing around the inhibitor-producing strain, and qualitatively, by assessing the clarity of the inhibition zone. Here we present the protocol for the deferred inhibition assay, describe ways to minimize variation between experiments, and define a clarity scale that can be used to qualitatively assess the degree of inhibition.
Introduction
Bacteria living in communities compete at the interspecific and intraspecific level for space and nutrients 6. Competition between microbes can often lead to exclusion of particular species or strains within a community. Exclusion can be due to competition for a particular receptor or nutrient, or the production of an antimicrobial compound by a member of the community 6.
Studies investigating competitive exclusion frequently examine the presence and absence of species using 16S rRNA sequencing 3,4,13. However, intraspecific trait variation, for example the presence or absence of antimicrobial peptide production, can hugely impact the ability of one species to inhibit another 1. Since 16S rRNA sequencing cannot distinguish between strains of the same species, intraspecific trait variation cannot be assessed. The technique described here can be used to assess the potential of individual strains to exclude other bacteria. This method has previously been used to show a positive correlation between the presence of Staphylococcus aureus inhibitory bacteria and the absence of S. aureus in a nasal community 7.
The deferred growth inhibition (competition) assay can identify production of antimicrobial compounds or nutrient competition by the inhibitor strain. It can also detect positive interactions, whereby the presence of the inhibitor strain actually improves the growth of the competitor strain. This assay can be used to create quantitative data (zone of clearing size) and qualitative data (degree of inhibition), both of which can be overlaid onto data of the presence or absence of a species within a community to find correlations between phenotype (trait) and competitive exclusion 7.
Protocol
The following method is adapted 7,8 to test competitor strains with inhibitor-producing strains.
NOTE: This protocol involves aerosol generation with bacterial cultures. This can only be performed in a contained space with implementation of locally-approved safety considerations. Spraying of the culture onto the agar plates within a level 2 safety cabinet will minimize contamination of the agar plates and the surrounding environment. This will also protect the researcher from aerosol inhalation of the sprayed bacteria, this is a major concern if the researcher is using containment level 2 organisms. Appropriate personal protective equipment must be worn. The area surrounding the petri dish is best covered with disposable absorbent tissue. Post-spray decontamination using UV light is advised.
1. Preparing the Agar Plates
- Prepare brain heart infusion (BHI) agar in accordance to the manufacturer's instructions.
- Autoclave the BHI agar at 121 °C, 15 psi for 20 min, or according to autoclave manufacturer's recommendations, to sterilize.
- Pour 15 ml aliquots of sterilized BHI agar into sterile petri dishes and allow it to set.
NOTE: Ensure every agar plate to be used within the assay originates from the same stock of agar and that there is the same amount of agar in each plate. This limits the variability in available nutrients and inhibitor diffusion between plates allowing comparability.
2. Preparing Sterile Broth
- Prepare BHI broth in accordance to the manufacturer's instructions.
- Add 10 ml aliquots of BHI to 28 ml glass universal bottles.
- Autoclave the BHI broth at 121 °C, 15 psi for 20 min, or according to autoclave manufacturer's recommendations, to sterilize.
NOTE: Ensure all broth used in the assay originates from the same batch of broth autoclaved at the same time to limit variation in nutrients which could cause variation in results.
3. Preparing Sterile Vaporizer Bottle(s)
- Dismantle the vapourizer bottle.
- Spray 5% surface active cleaning agent through the vapourizer to eliminate internal contaminants, soak the vapourizer bottle, lid and vapourizer in the 5% surface active cleaning agent overnight.
NOTE: Gloves and appropriate personal protective equipment should be used when handling surface active cleaning agent. - Dry the vaporizer, lid and bottles under sterile air flow hood.
- Fill the bottle with 70% ethanol, reattach vapourizer and spray some of the ethanol through the vapourizer.
- Put the lid on the vaporizer and store with the ethanol inside until use.
- Immediately prior to use, aseptically rinse the bottle with sterile BHI broth and spray the broth through the vapourizer.
NOTE: This prevents the ethanol affecting the inoculum added to the bottle in section 6.3.
4. Preparing the Bacterial Overnight Culture(s)
- Streak the competitor and inhibitor strains onto a sterile BHI agar plate and incubate at 37 °C aerobically for approximately 18 hr (overnight).
NOTE: This plate produces a stock of bacteria that can be used for multiple replicates within the same week when stored at 4 °C. - Inoculate 10 ml of sterile BHI broth with a single colony of the required competitor bacterial strain within a 28 ml universal bottle.
- Incubate the universal bottle at 37 °C aerobically overnight by shaking at 250 rpm in an orbital shaker.
5. Spot Culture of the Inhibitor-producing Isolate
- Prepare the overnight culture(s) as described in section 4.
- Ensure agar plates are completely dry, with no condensation on the lid, prior to inoculation (e.g. by incubating plate for 60 min at 37 °C); this prevents the inoculum from spreading across the plate from the inoculation site.
- Pipette 25 µl of the overnight culture onto the center of the agar plates. Avoid completely depressing the pipette plunger to prevent air bubbles that spray culture across the agar.
- Allow the culture to dry at room temperature to limit spread of the culture. Incubate the agar plates at 37 °C aerobically overnight.
6. Preparing the Spray Inoculum of the Test Competitor Strain(s)
- Prepare the overnight culture as described in section 4. Dilute the overnight culture 10 fold in BHI broth to yield approximately 4 x 106 CFU ml-1, and a suspension of OD600 0.3 ±0.05.
NOTE: A 10 fold dilution will not give 4 x 106 CFU ml-1 for all bacterial species, calculate the necessary dilution factor by spread plating serial dilutions between 1 x 10-1-1 x 10-6. - Pour the diluted culture into a sterile plastic perfume vaporizer bottle.
NOTE: The total volume must be more than is necessary for the number of agar plates being sprayed. Approximately 250 µl per plate is suitable for a 9 cm diameter Petri dish.
7. Deferred Growth Inhibition Assay
- Spray the culture through each of the vaporizer bottles to ensure that the culture is loaded in the spray mechanism prior to spraying onto the agar.
- From a distance of approximately 15 cm above the agar, spray approximately 250 µl of diluted culture (3 sprays of a typical 50 ml vaporizer) over the entire agar surface. Incubate the agar plates at 37 °C aerobically overnight. Repeat for other plates with the same number of sprays.
8. Interpreting Deferred Growth Inhibition Assay Results
- Measure the growth inhibition zone of the sprayed competitor strain (Figure 1A–C, "X" to "X") in millimeters, subtracting the diameter of the central spot of the inhibitor-producer (Figure 1A–C, "Y" to "Y").
NOTE: Figure 1E shows how this data could be presented - Record the clarity score for zones of inhibition.
NOTE: This indicates the degree of inhibition observed. Examples are shown in Figure 1A–D, strains that can be used for standardization are suggested in the representative results section. The following scale can be used for clarity scoring: a clarity score of 0 to describe a zone of complete inhibition of the competitor strain's growth. A score of 1 to describe a zone of almost complete inhibition of growth, with only individual colonies present in the zone of clearing. A score of 2 to describe a visible zone of reduced growth, growth within this zone is confluent or near confluent but reduced in density by >50 %. A score of 3 to describe only minimal growth reduction, growth is confluent and reduced by less than 50 %, a zone is still visible and measurable. A score of 4 to describe no visible inhibition of growth. A score of 5 to describe that the growth of the competitor strain was increased in the proximity of the test inhibitor-producing strain. See Figure 1 for examples. These scores are subjective to the experimenter.
9. Replicates
- Repeat assays at least in biological triplicate to determine the reproducibility of the results obtained.
NOTE: If replicates are performed on different days, take measurements and score plates immediately after removal from incubation, then either store plates at 4 °C or take high quality photographs of plates before discarding them. Storage or photographs of plates enables easy comparison of results between different days, this is particularly important for confirming reproducible scoring across days/experiments. Be aware that storage of the plates has the potential to alter results.
NOTE: For clear photographs place plates upright without their lid on a matte black background in an area of diffused light, use a high resolution camera (≥8 MP) capable of focusing on objects close to the lens (macro mode). Alternatively, some gel imagers with trans-illuminator set to white light can produce high quality, highly reproducible images.
Representative Results
The assay results should be observable as a difference in the overlaid competitor strain near the centrally spotted inhibitor-producing strain (Figure 1). These differences can be interpreted as full inhibition (Figure 1A), partial inhibition (Figure 1B and 1C), no inhibition (Figure 1D) or positive association. The widely available lab strains S. epidermidis Tü3298 and S. epidermidis Rp62A, and S. epidermidis Rp62A and S. aureus Newman were used in Figures 1B and 1D respectively. We suggest that these strains could be used to test this protocol.
Depending on species tested, full inhibition and partial inhibition are most likely the result of antimicrobial peptides, antibiotics or other diffusible growth inhibitors produced by the inhibitor-producing strain. Partial inhibition has also been linked to reduced nutrient availability for the competitor strain due to uptake or sequestering of nutrients by the inhibitor strain 6.
Comparison of the size of the zone of inhibition between different inhibitor-producing isolates tested against the same competitor strain indicates differences in any of the following: the amount of antimicrobial produced by the inhibitor-producing strain; the ability of the inhibitor to diffuse through the media; the type of inhibitor produced and the resistance level of the competitor strain to the inhibitors being produced. Comparison of the size of the zone of inhibition between different competitor strains tested against the same inhibitor-producing strain is indicative of the resistance levels of the competitor strains to the inhibitor being produced.
Results will vary depending on the inhibitory strain used and the competitor strain used. Repeats using the same inhibitory and competitor strain should ideally have the same clarity score (described in section 8.2), this indicates good technique has been used throughout.
If inconsistent application of bacteria has occurred the plates should not be used. For example, if the test competitor culture is too dilute the zone of inhibition will appear larger and with a less clearly defined edge than is typically observed (Figure 2A); this would affect reliability of any comparative zone measurements. If the inhibitor-producing strain spot was not fully dried before incubation overnight, the inhibitor strain is likely to spread and not form a clearly defined spot (Figure 2B), which typically affects zone measurements but can also affect the clarity scoring. If the competitor strain is not sprayed evenly this can lead to an uneven spread of bacteria (Figure 2C). If the competitor strain is too concentrated it is possible that the competitor strain may grow on top of the inhibitor strain (Figure 2C), in extreme cases this can lead to some inversion of competitor/inhibitor strain roles.
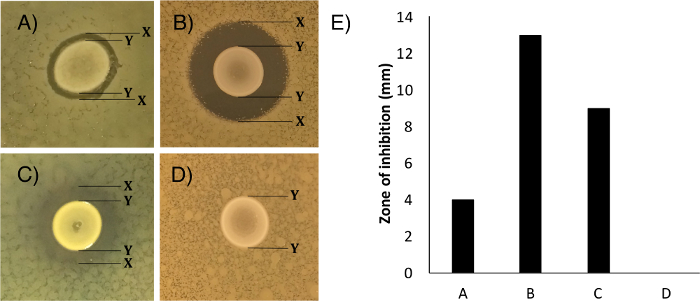
Figure 1: The range of qualitative and quantitative results that can be obtained from the deferred growth inhibition assay. Results for the growth inhibition can range from (A) complete inhibition (score of 0), (B) partial inhibition (score of 1), (C) partial inhibition (score of 2) to (D) no inhibition (score of 4). Labels indicate the edges of growth of the inhibitor strains (Y) and the outermost edge of the inhibition zones (X). The size of the zone of inhibition range from 0 mm to more than 10 mm, as shown in (E), which shows the measurements for the plates photographed in A, B, C and D. Results are shown for recent staphylococcal isolates (A, C) or common lab staphylococcal isolates S. epidermidis Tü3298 (inhibitor) and S. epidermidis Rp62A (competitor) (B), and S. epidermidis Rp62A (inhibitor) and S. aureus Newman (competitor) (D). Please click here to view a larger version of this figure.
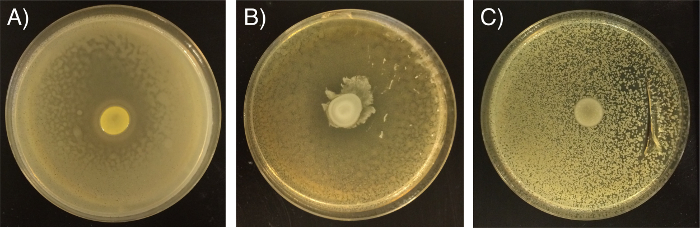
Figure 2: Examples of inconsistent results due to poor technique. Results may appear different than expected if (A) competitor strain was too dilute, (B) inhibitor strain was not fully dry before incubation/plates were not dry or (C) if the competitor strain was not sprayed evenly and was too concentrated. Please click here to view a larger version of this figure.
Discussion
A critical step of this bacterial species competition protocol that differentiates it from others, is the overnight incubation of the inhibitor isolate before addition of the competitor isolate. The inhibitor strain requires this growth period to produce inhibitory compounds. As the competitor strain is overlaid on top of the same medium, this method allows the researcher to observe the full potential of the inhibitor strain to interfere with growth of the competitor strain. As a result, this assay reflects what happens with nature; one isolate is fully established in a niche, and the other must be able to counteract any inhibitory variables to be able to successfully invade the niche.
There are two parts to this protocol that are most likely to cause erroneous results if performed incorrectly. Firstly, incomplete sterilization of the vaporizer bottles can introduce contaminating bacteria into the assay. Incubation of the broth used to rinse the vaporizer bottle in step 3.6 in a sterile vessel can be used to confirm bottles were completely sterilized. Additional washes in disinfectant solutions, such as Virkon, can be used prior to the wash in surface active cleaning agent, described in step 3.2, if additional sterilization is required.
A second step that may cause erroneous results is the dilution of the competitor strain (step 6.1). If the dilution is not optimal, clear measurements of inhibition may not be easily determined. This assay was optimized using various nasal isolates as inhibitor-producing strains across a wide range of Gram-positive and Gram-negative species versus S. aureus SH1000 as the competitor strain 7,8. It has subsequently been tested using various coagulase-negative and coagulase-positive staphylococci as competitor and inhibitor-producing strains. When testing genera other than those described above as the competitor strain, some optimization of the dilution for the inoculum may be necessary.
If clarity scores vary significantly between repeats, it might be necessary to standardize the competitor strain inoculum by setting it to a specific optical density. However, this will increase the time the assay takes to set up, and is likely to reduce the number of strains it is possible to process per experiment. Visually adjusting the culture to an appropriate OD using a McFarland standard may provide a rapid alternative.
One limitation of this technique is that it it can only be applied to culturable bacteria. This is why many studies use 16S rRNA sequencing to predict competitive exclusion interactions 3,4,13. However, genetic and metagenomic techniques can only distinguish community members to the species level, and/or do not account for particular intraspecific trait variation. It is possible to screen for the association of a particular gene that encodes a trait with the presence or absence of a bacterial species 5, however this requires pre-knowledge of a gene likely to be associated with exclusion of a species.
A safety consideration of this technique is that, due to the aerosol generation of bacteria, it can only be used in laboratories with a level 2 safety cabinet, or with similar safety precautions. There are alternative techniques to test for competition between isolates that account for intraspecific trait variation and do not generate aerosols of bacteria. One such method is the simultaneous antagonism assay 2. In this method, an inoculum of one isolate is spread onto an agar plate, and a second isolate inoculum is spotted or stabbed on top once the first has dried. This would produce an environment where the strains are in direct competition (simultaneous antagonism), both competing to produce inhibitory compounds and scavenge nutrients first. The advantage of the deferred growth inhibition assay over the simultaneous antagonism assay is that the inhibitor isolate should always have the competitive advantage of being fully established before invasion of the competitor isolate. This is more reflective of what would happen in the environment, as in most niches, one isolate will be established before another strain is added, rather than two isolates being added at the same time 8.
Another alternative to avoid aerosol generation of bacteria would be to add the competitor isolate in top agar, which could be poured rather than sprayed over the inhibitor isolate. However, the volume of top agar added would need to be approximately 3 ml to ensure even coverage of the plate. Within this volume, the competitor isolate would not be growing on the same media as the inhibitor isolate, so would not suffer from reduced nutrients. The competitor would also not be in direct contact with any inhibitory compounds until they diffused into the top agar. Such an assay could not be described as deferred inhibition or simultaneous antagonism.
A similar method has previously been used to assess the levels of bacitracin resistance amongst clinical S. aureus isolates 10. The main difference between the protocol described here and that described by 10, is the latter only classified competitor strains as sensitive (complete inhibition- clarity score 0) or resistant (partial to no inhibition- clarity score 1-5). The deferred growth inhibition assay could therefore also be used to screen for levels of resistance to producers of particular antimicrobials, or even to search for novel antimicrobials active against multi-drug resistant pathogens.
It has been suggested that the host microflora could be manipulated to eliminate undesirable members of the bacterial community 9. Application of corynebacteria or Staphylococcus epidermidis have already been shown to eliminate S. aureus colonization in the nares in a significant proportion of the population 11,12. Studies into specific strains that can competitively exclude undesirable members of the host flora, through techniques such as the one described here, may therefore lead to future therapeutics.
Disclosures
The authors have nothing to disclose.
Acknowledgements
JCM is funded by a BBSRC-IPA award with Unilever Plc (BB/L023040/1). HAG is funded by a Saudi Arabian Ministry of Higher education PhD studentship.
Materials
| Brain heart infusion broth | Lab M | lab049 | rich general purpose media |
| agar-agar | Merck Milipore | 101614 | |
| petri dishes | Sarstedt | 82.1473.001 | |
| plastic perfume vaporizer | not known | – | 10 cm high with 3.5 cm diameter clear plastic spray bottle with lid, purchased from the travel section of a supermarket |
| Universal bottles | no longer in production, alternative SLS BOT5006 | 28 ml glass cylindrical bottle (approx 280 mm diameter, 850 mm height), flat bottomed with plastic screw cap | |
| Decon 90 | Decon | surface active cleaning agent |
References
- Bolnick, D. I., et al. Why intraspecific trait variation matters in community ecology. Trends Ecol Evol. 26 (4), 183-192 (2011).
- Christensen, G. J. M., et al. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genomics. 17 (152), (2016).
- Faust, K., et al. Microbial co-occurrence relationships in the human microbiome. PloS Comput Biol. 8 (7), (2012).
- Frank, D. N., Feazel, L. M., Bessesen, M. T., Price, C. S., Janoff, E. N., Pace, N. R. The Human nasal microbiota and Staphylococcus aureus carriage. PLoS One. 5 (5), e10598 (2010).
- Fredheim, E. G., Flaegstad, T., Askarian, F., Klingenberg, C. Colonisation and interaction between S. epidermidis and S. aureus in the nose and throat of healthy adolescents. Eur J Clin Microbiol Infect Dis. 34, 123-129 (2015).
- Hibbing, M. E., Fuqua, C., Parsek, M. R., Peterson, S. B. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 8, 15-25 (2010).
- Libberton, B., Coates, R. E., Brockhurst, M. A., Horsburgh, M. J. Evidence that intraspecific trait variation among nasal bacteria shapes the distribution of Staphylococcus aureus. Infect Immun. 82 (9), 3811-3815 (2014).
- Libberton, B., Horsburgh, M. J., Brockhurst, M. A. The effects of spatial structure, frequency dependence and resistance evolution on the dynamics of toxin-mediated microbial invasions. Evol appl. 8, 738-750 (2015).
- Liu, C. M., et al. Staphylococcus aureus and the ecology of the nasal microbiome. Sci Adv. 1, e1400216 (2015).
- Nascimento, J. S., Ceotto, H., Nascimento, S. B., Giambiagi-deMarval, M., Santos, K. R., Bastos, M. C. Bacteriocins are alternative agents for control of multiresistant staphylococcal strains. Lett Appl Microbiol. 42, 215-221 (2006).
- Park, B., Iwase, T., Liu, G. Y. Intranasal application of S. epidermidis prevents colonization by methicillin-resistant Staphylococcus aureus in mice. PLoS One. 6 (10), e25880 (2011).
- Uehara, Y., et al. Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J Hosp Infect. 44, 127-133 (2000).
- Wos-Oxley, M. L., et al. A poke into the diversity and associations within human anterior nare microbial communities. ISME J. 4, 839-851 (2010).

