Immunostaining of Biocytin-filled and Processed Sections for Neurochemical Markers
Summary
This protocol presents a method for the morphological recovery of neurons patched during electrophysiological recordings using biocytin filling and subsequent immunohistochemical postprocessing. We show that thick biocytin-filled sections that were stained and coverslipped can be restained with a second primary antibody days or months later.
Abstract
Electrophysiological recordings of cells using the patch clamp technique have allowed for the identification of different neuronal types based on firing patterns. The inclusion of biocytin/neurobiotin in the recording electrode permits post-hoc recovery of morphological details, which are necessary to determine the dendritic arborization and the regions targeted by the axons of the recorded neurons. However, given the presence of morphologically similar neurons with distinct neurochemical identities and functions, immunohistochemical staining for cell-type-specific proteins is essential to definitively identify neurons. To maintain network connectivity, brain sections for physiological recordings are prepared at a thickness of 300 µm or greater. However, this thickness often hinders immunohistological postprocessing due to issues with antibody penetration, necessitating the resectioning of the tissue. Resectioning of slices is a challenging art, often resulting in the loss of tissue and morphology of the cells from which electrophysiological data was obtained, rendering the data unusable. Since recovery of morphology would limit data loss and guide in the selection of neuronal markers, we have adopted a strategy of recovering cell morphology first, followed by secondary immunostaining. We introduce a practical approach to biocytin filling during physiological recordings and subsequent serial immunostaining for the recovery of morphology, followed by the restaining of sections to determine the neurochemical identity. We report that sections that were filled with biocytin, fixed with paraformaldehyde (PFA), stained, and coverslipped can be removed and restained with a second primary antibody days later. This restaining involves the removal of the coverslip, the washing of sections in a buffer solution, and the incubation of primary and secondary antibodies to reveal the neurochemical identity. The method is advantageous for eliminating data loss due to an inability to recover morphology and for narrowing down the neurochemical markers to be tested based on morphology.
Introduction
The brain is known for diversity in the structural and functional characteristics of its individual neuronal elements. Understanding the roles of distinct neuronal types in brain function and pathology requires characterization and unambiguous identification of neurons. Structurally, the morphological features defined by somato-dendritic location determine the potential inputs that a given neuron receives, while the pattern of axonal arborization identifies potential postsynaptic targets. The structural diversity of neurons has been appreciated since the days of Ramón y Cajal's seminal histological studies1. The advent of single-cell recording techniques revealed that structurally distinct neurons also show differences in firing patterns and synaptic characteristics. The diversity in structure and physiology is particularly evident in GABAergic inhibitory neurons2,3. In addition, it has become increasingly apparent that structurally similar neurons can express different neurochemical markers and show corresponding functional differences4. Similarly, neurons with the same neurochemical markers can have distinct structures and functions5-10. Thus, in practice, the analysis of the functional characteristics of neurons and their role in the network entails defining both the morphological and neurochemical identities. Even with the advent of reporter mouse lines targeting specific neurochemical markers, it is often necessary to determine morphology and subtype identity based on immunohistology11.
The standard method used to characterize cells recorded in acute brain slices is to fill them with biocytin or neurobiotin during the recording, fix the sections in paraformaldehyde (PFA) following the recordings, and use immunohistochemistry to reveal the morphology and neurochemistry. Since the thickness of sections for slice physiology are typically 300 µm or more, and because most antibodies fail to penetrate all the way through that depth, the slices need to be re-sectioned to 60 µm or less to allow for simultaneous immunostaining for biocytin and neurochemical markers12-14. Unfortunately, resectioning is laborious; risks loss of tissue during sectioning; and can lead to differential tissue shrinkage, complicating morphological reconstructions. Additionally, prior knowledge of morphology could help narrow down the candidate markers that are likely to be expressed by the cells. We have modified the standard biocytin immunohistology protocols to allow serial processing of sections first for the recovery of morphology and then for the identification of potential neurochemical markers.
Immunohistochemistry is the study of antigen distribution in tissues or cells and can be visualized using an enzyme, fluorescent labels, radioactive elements, or gold colloid particles15. The procedure involves using primary antibodies to specifically tag and amplify one or more specific antigens, followed by the use of fluorescent secondary antibodies targeting the primary antibody for visualization. Due to the need to distinguish the fluorescence spectra of each secondary antibody without overlap, only a limited number of antigens can be examined simultaneously. Thus, prior knowledge of morphology could be useful in selecting the candidate neurochemical markers for cell classification. Conceptually, the rationale behind serial processing of already-stained sections is based on the premise that immunolabeling for one protein or peptide should not interfere with antigenicity and subsequent immunolabeling for a structurally independent peptide16. This lack of interference is due to the binding of the antibodies to a specific protein epitope on an antigen and therefore allows for the simultaneous staining of multiple antigens in the same tissue. The number of antigens revealed by immunostaining is limited by the need for non-overlapping spectra of the fluorescent secondary antibodies and by the need to target individual antigens with antibodies raised in different species so as to eliminate cross-reactivity17,18. While this is the reasoning behind serial rather than simultaneous labeling with two distinct antibodies that may interact, to our knowledge, immunostaining for a second antigen has not been reported after the completion of immunolabeling for one or more antigens on mounted sections. Here, we describe a method for serial immunostaining of previously stained and mounted sections. While we detail this process for a serial immunolabeling procedure for the recovery of morphology followed by staining for protein/peptide markers in thick sections, the same procedures can be used in standard, thin histological sections as well. In addition, we describe a practical approach to fill recorded neurons with biocytin and the process to dislodge the electrode from the cell upon completion of recordings to optimize the filling of the axonal and dendritic arbors of neurons, as presented in our recent work6,8.
The most crucial advantage of the procedure described here is that the morphology of the recorded cell can be fully recovered and imaged before attempting to resection or immunostain the slices. Although issues with the penetration of certain antibodies may render it necessary to resection slices for secondary immunostaining, the procedures detailed here would eliminate the need to reconstruct complex neurons from multiple sections and would avoid issues due to tissue loss and differential shrinkage, which can compromise reconstruction following resectioning. An added advantage is that the process will reduce cost, time, effort, and expensive antibodies by limiting immunostaining and re-sectioning to slices in which biocytin-filled neurons are recovered. The most practical aspect is the additional immunostaining that can be performed on sections stained months before using the aforementioned technique. In particular, the recovery of morphology would considerably reduce the potential that physiological data from the cells is discarded due to an inability to obtain a basic morphological characterization of the cell type.
Protocol
1. Biocytin Filling during Electrophysiology
NOTE: The readers can refer to alternative sources for basic patch-clamp recording techniques and instrumentation19-22, which are not elaborated upon here. The steps detailed here assume that the equipment and procedures for patch-clamp recordings are already established, and the description will be restricted to details related to biocytin-filling and post-hoc immunostaining. All experiments outlined in this manuscript were performed on rats.
- Using a vibratome, prepare live brain sections of the desired region at a thickness of 300 – 350 µm23.
- Prepare an internal solution containing 0.2% biocytin by adding 2 mg of biocytin to 1 mL of internal solution6. For best results, sonicate the internal solution for 10 – 15 min until the biocytin dissolves completely. Next, load the internal solution in a 1 mL syringe fitted with a 0.2 µm pore size polypropylene filter tip attached to a microloader. Keep this loading syringe on a cold pack to maintain the stability of the Mg-ATP and Na-GTP in the internal solution.
NOTE: An internal solution containing potassium gluconate (126 mM K-gluconate, 4 mM KCl, 10 mM HEPES, 4 mM Mg-ATP, 0.3 mM Na-GTP, and 10 mM phosphocreatine) is optimal for the recovery of certain neurochemical markers, including parvalbumin, during immunolabeling. In our experience, using cesium- and chloride-based internal solutions reduces the ability to recover immunostaining for certain neurochemical markers. Neurobiotin or a cell-impermeant, fixable, polar tracer combining an Alexa Fluorophore with biocytin can be used as an alternative to biocytin. - Under infrared differential interference contrast, visualize the appropriate cell for recording. To minimize any disruption of the axon, approach the cell body by lowering the pipette along its axis using the "approach" function available in most micromanipulators. Establish whole-cell recordings under infrared differential interference contrast using patch-clamp physiology protocols19-22.
NOTE: Avoid approaching the cell from the direction of the putative axon, as it will sever the axon, visualized as a bleb at the cut end (green arrow in Figure 1). Also, avoid applying high positive pressure to the recording pipette while approaching the cell to minimize the spill of biocytin, which will result in high background staining.
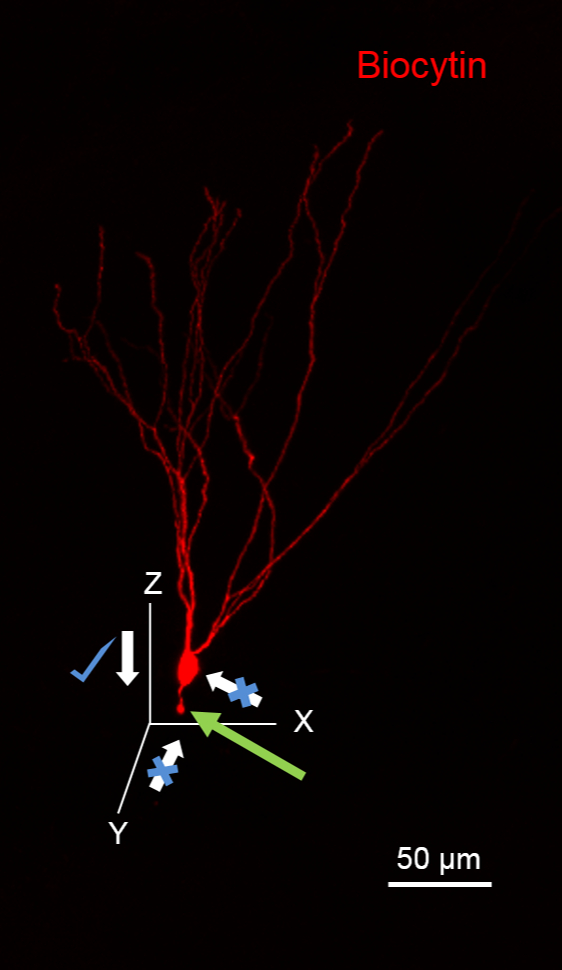
Figure 1. Suggested Plane for Approaching Cells for Biocytin Fills. The image illustrates a Granule Cell (GC) fill with biocytin during recording that has been processed to reveal biocytin (in red using 594-conjugated streptavidin). The GC has its dendrites oriented in the XY plane. Notice the severed granule cell axon (green arrow) due to pipette movement along the XY plane. Note that it is ideal to approach this neuron along the XZ plane. Scale bar: 50 µm.
- In voltage clamp configuration, determine the whole-cell capacitance and series resistance in response to small (5 mV), brief (30 ms) voltage steps using the using the membrane test function in bath mode in an electrophysiology data acquisition and analysis software. This will ensure the establishment of whole-cell recording conditions to allow for biocytin-filling.
- Conduct physiological recordings in either current- or voltage-clamp mode as needed. A minimum recording time of >10 min at 34 °C is ideal.
NOTE: Longer recording times may be needed for studies performed at room temperature. - Upon completion of the physiological recordings, re-establish the patch-clamp configuration by slowly moving the recording pipette in small steps, alternating upward (along the Z-axis) and outward (along the X-axis) in voltage-clamp mode.
- Simultaneously monitor the capacitance and input resistance using the "membrane test" function to visualize the loss of capacitive transients and the collapse of the current responses to a straight line, indicating the re-sealing of the cell and the establishment of an outside-out-patch at the pipette tip. Holding the cell at a depolarized potential (-40 mV) will facilitate the re-sealing process.
NOTE: This procedure for the removal of a cell typically ensures the complete recovery of the soma and dendrites. Do not apply positive pressure to the recording pipette during this procedure or while detaching the cell from the pipette. Visualization of the soma of the recorded cell in the slice indicates successful disengagement. The presence of cellular debris or a membrane patch at the end of the detached tip typically indicates dislocation of the soma and will not yield complete cellular morphology. - After detaching the pipette from the cell, retain the slice in the recording chamber for 3 – 5 min to ensure the transport of the dye to distal dendritic and axonal processes.
- Transfer the slices to a 24-well plate containing 4% PFA for fixation.
CAUTION: PFA is carcinogenic, and appropriate personal protective equipment, including gloves and a face mask, must be used to avoid skin irritation and inhalation. PFA is flammable and must be kept out of reach of fire. PFA is never disposed of in the drain and must be collected as chemical waste. PFA fixation must be isolated from areas utilized for live slice preparation to avoid contamination of the physiology setup, including transfer pipettes. - 24 – 48 h after PFA fixation, transfer the slices to 0.1 M Phosphate-buffered Saline (PBS) for storage prior to immunostaining.
NOTE: Ideally, biocytin recovery and immunostaining are best when performed soon after fixation, although staining performed within 7 d of fixation yields good results. Slices can be maintained in PBS with 0.02 – 0.05% sodium azide for staining performed up to and over 90 days after fixation. Although overnight fixation in PFA works well for most antibodies, it is possible that some antibodies work best when the duration of incubation in fixative is reduced. Fixation in PFA for over 48 h reduces the availability of antigens and diminishes the chances of successful secondary immunostaining for neurochemical markers.
CAUTION: Sodium azide is extremely hazardous and an irritant upon contact with the skin or eyes or upon ingestion or inhalation. Severe over-exposure can result in death. The carcinogenicity and mutagenicity of the compound are not known. Use appropriate personal protective equipment including gloves, splash goggles, a lab coat, and a dust respirator. Sodium azide is never disposed of in the drain and must be collected as chemical waste.
2. Staining of the First Primary Antibody and Biocytin
(Day 1)
NOTE: The following immunostaining procedure is for free-floating sections and requires continuous shaking at a low speed (2 revolutions/min, rpm) on a shaker for all incubation steps.
- Wash the tissue 3x for 10 min each in 0.1 M PBS (pH = 7.4).
NOTE: 0.1 M PBS can be commercially purchased or made in the laboratory as follows (makes 1 L): 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 in 1 L of dH20.
NOTE: If the desired neurochemical marker is known, staining for a protein/peptide can be performed simultaneously with biocytin staining (steps 2.2 – 2.4). If staining for biocytin only, proceed to step 2.5. - OPTIONAL: Block with 10% Blocking Serum (BS; normal goat serum (NGS) diluted in 0.3% Triton X-100 in PBS for 1 h at RT).
NOTE: Each well should receive the same volume of the solution; the recommended volume is between 250 – 500 µL/well. The selection of the blocking serum is based on the species in which the secondary antibodies are raised (e.g., NGS is used for dye-conjugated goat anti-(respective primary antibody species)). If the need arises to use secondary antibodies raised in different species to immunostain sections, include 10% of each normal serum in blocking step (2) and 3% of each normal serum for the primary and secondary antibody incubation steps. - (OPTIONAL) Incubate the sections in primary antibody, CB1R (polyclonal, guinea pig) with 0.3% Triton X-100, and 3% BS in 0.1 M PBS at RT O/N. CB1R is used to identify cannabinoid receptor type 1 expression.
NOTE: This step is optional and not required to reveal the biocytin filling. Figure 2 illustrates a section labeled for biocytin and CB1R during the first staining process. O/N incubation at RT is sufficient for most of the primary antibodies; however, certain primary antibodies, including CB1R, require 3 – 5 d of incubation. If the incubation needs to be longer than 1 d, it is recommended to incubate at 4 °C because Triton X-100 can permeabilize slices and can lead to the disintegration of the tissue during prolonged incubation at room temperature. Include a negative control lacking primary antibody in at least one section for each series of experiments as a control for the specificity of the secondary antibody. For negative controls, the primary antibody is omitted, but the 0.3% Triton X-100 in 0.1 M PBS and BS are added.
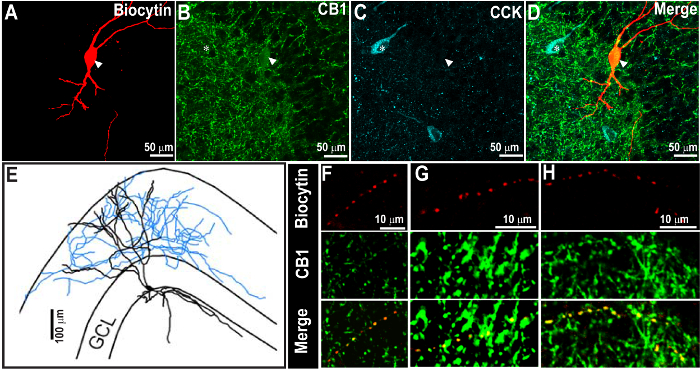
Figure 2. Successful CCK Staining 1 Week Following the Recovery of Biocytin Staining and Cannabinoid Receptor Type 1 (CB1R)-labeling. (A – C) Confocal images at 60X showing the biocytin-filled neuron (A) and CB1R immunoreactivity (B), indicated by the arrowhead. The same section was processed for CCK immunostaining after 1 week (C). The overlay of images is shown in D. CCK immunoreactivity was revealed using far-red and was pseudo-colored in cyan. (E) Morphological reconstruction of the biocytin-filled cell in A. Note that the biocytin and CB1R staining are evident even after the second CCK immunostaining. Also note the expected distribution of CB1R and CCK immunostaining patterns. (F – H) Magnified image of the axon from the cell, as in A, show close co-localization of biocytin and CB1R in axons (arrow heads). Scale bar: 50 µm (A – D), 100 µm (E), and 10 µm (F – H). Images are reproduced from Yu et al. 20159. Please click here to view a larger version of this figure.
(Day 2)
- Wash the brain sections 3x for 10 min each in 0.1 M PBS (pH = 7.4).
- Incubate the sections in streptavidin red dye conjugate (concentration: 1:1,000; excitation: 594 nm with emission in the visible red spectrum) with 0.3% Triton X-100 and 3% BS in 0.1 M PBS at 4 °C O/N in the dark (wrapped in aluminum foil) to reveal the biocytin. OPTIONAL: Include a secondary antibody, goat anti-guinea pig (concentration: 1:500; excitation: 488 nm and emission in green), with the above incubation if following the optional primary incubation in step 2.3.
NOTE: Secondary antibody will be needed only if the optional primary antibody staining (step 2.3) is performed. To visualize biocytin, a streptavidin-fluorophore conjugate with an emission spectrum in red is recommended, as it helps to visualize finer axons in detail and with better contrast (Figures 1 – 3). During double or triple immunostaining, in order to avoid cross-reactivity of secondary antibodies with each other, add secondary antibodies one after the other in a serial fashion (2 h interval between serial additions of the antibodies is sufficient). Figure 3A illustrates a biocytin-filled cell processed without the optional primary antibody staining and imaged prior to resectioning.
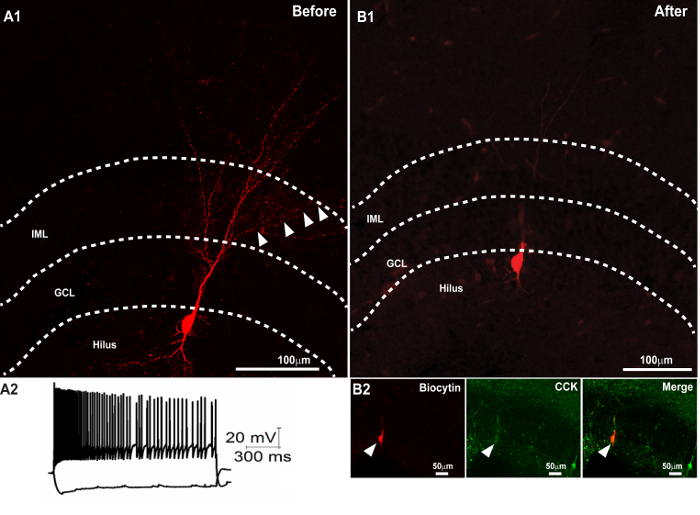
Figure 3. Successful Second Immunostaining following the Recovery of Biocytin Morphology and Resectioning. (A1) Confocal image of a 300 µm slice showing the recovery of the dendritic morphology of a biocytin-filled cell. (A2) Membrane voltage traces of the neuron in response to +500 and -100 pA current injections. (B) Recovery of the biocytin-filled soma in a 50 µm section obtained after resectioning the 300 µm section (in A1) after mounting and imaging. (B2) Subsequent immunostaining of the thin section revealed colocalization of the biocytin-filled soma (right panel) with CCK (middle panel). The merged image is illustrated in the right panel. Scale bar: 50 µm. Panel A2 is reproduced with permission from Yu et al., in press25. Please click here to view a larger version of this figure.
(Day 3)
- Wash brain sections 3x for 10 min each in 0.1 M PBS (pH = 7.4).
- To protect the fluorescence and to facilitate re-staining in the future, mount the sections in an aqueous-based mounting medium and seal the edges of the coverslip with clear nail polish.
NOTE: It is best to restain sections as early as possible to avoid difficulties in removing the nail polish from the slide, mold infestation, and dehydration of tissue due to leakage of the mounting medium from the glass slide. Microbial contamination can be prevented by using 0.02% sodium azide in PBS.
CAUTION: Sodium azide is highly toxic and flammable when it is in contact with water. Please refer to standard operating procedures for the handling and disposal of hazardous chemicals.
(Days 4 – 7) - Perform confocal imaging with excitation at 594 and 488 nm and at a magnification of 20X or 40X to reveal biocytin-filled neurons in red and neurochemical markers (CB1R) in green. Assess colabeling (Figure 2).
- Set the camera exposure to less than 100 ms to avoid photobleaching and obtain confocal image stacks of the axonal and dendritic arbors of the entire neuron. Use confocal image stacks and neuron tracing software packages to reconstruct the neuronal morphology.
3. Staining of the Second Primary Antibody
NOTE: While the second immunostaining step can be performed 90 d after the first staining, optimal staining is achieved if the second primary antibody staining is performed within 7 – 10 d.
(After 7 – 90 d)
- Apply acetone to a cotton swab and strip the nail polish off of the glass slide.
- Remove the coverslip carefully using fine-tip forceps and put a few droplets of 0.1 M PBS on the sections.
- Carefully remove the sections from the slide using a fine paint brush and wash them in 0.1 M PBS for 24 – 48 h on a shaker covered in aluminum foil in low light at 4 oC.
NOTE: It is crucial to cover sections with aluminum foil to avoid photobleaching. - To ensure complete removal of the mounting medium, wash the sections 1 – 2x on the following day in 0.1 M PBS for 10 min each.
- Proceed with incubation in the second primary antibody (e.g., cholecystokinin, a neuropeptide found in certain GABAergic interneurons (CCK; concentration: 1:1,000; mouse monoclonal antibody)) for 1 d (Figure 2).
NOTE: This procedure is similar to step 2.3 but uses a different antibody. Additionally, the blocking step is omitted during re-staining. - Wash the brain sections three times for 10 min each in 0.1 M PBS (pH = 7.4).
- Stain with the fluorophore-conjugated secondary antibody goat anti-mouse (concentration: 1:500; excitation wavelength: 647 nm), making sure that the excitation-emission spectra of the secondary antibody do not overlap with streptavidin (concentration: 1:1,000; excitation wavelength: 594 nm) and prior immunofluorescence stains.
- Mount the sections in aqueous mounting medium and seal the edges of the cover slip with clear nail polish.
NOTE: Store the sections at 4 °C in an opaque storage box to protect the fluorescence.
Representative Results
Upon successful completion, the sections retain the biocytin fill and the immunolabeling performed in step 2 and can be imaged using confocal or epifluorescence microscopy. In addition, the processed sections will also show immunostaining for the antigen labeled during the subsequent processing in step 3. In the section illustrated in Figure 2, the morphology of a biocytin-filled neuron in a thick section (300 µm) was revealed using streptavidin visualized in red and the first primary (CB1R) visualized in green. The first immunostaining was performed within 7 d of PFA fixation. The slice was mounted in an aqueous mounting medium, imaged using a confocal microscope, and stored for a period of two months before re-staining for the CCK antibody. Note the morphology of the recorded neuron, including axonal arbors (Figure 2E), was reconstructed from the confocal images obtained after the first staining procedure. Colocalization of the axon with CB1R immunoreactivity (Figure 2 F – H) was expected based upon physiological studies. The second staining for CCK was performed on sections two months after the first staining procedure. CCK was imaged in far-red and was pseudo-colored in cyan for visualization. Similarly, Figure 3 illustrates an example of a cell in which biocytin-labeling was revealed and imaged in a thick section (Figure 3A1). The intrinsic physiology of the neuron is illustrated in Figure 3A2. The slice was re-sectioned at 50 µm and, although some sections containing dendritic processes were lost, the section containing the soma recovered (Figure 3B1). Subsequent CCK immunostaining of the thin section showed CCK expression in green in the biocytin-labeled soma. Mounting the thick slice in an aqueous medium prevents excessive tissue shrinkage. On average, 300-µm sections mounted using the aqueous medium shrink by about 25.67 ± 3.14% (n = 5 slices) when measured one week after mounting. Thus, the thick sections mounted using the aqueous medium are ~225 µm, allowing for resectioning. In several control studies, it was confirmed that the process of immunolabeling for the second time did not lead to non-specific labeling by the antibody. For example, it is known that parvalbumin (PV) and CCK immunoreactivity are mutually exclusive in hippocampal neurons. Controls in which the first immunolabeling step (2) for biocytin (red) and CCK (green) was followed by the second labeling for parvalbumin (with excitation at 405 nm and emission in blue) showed non-overlapping expression patterns (Figure 4). Additionally, the distinctive laminar axonal staining patterns of the neurons expressing specific neurochemical markers (PV, CCK, CB1R) were not altered by the serial re-staining procedures (Figures 2 – 4). Moreover, secondary staining consistently revealed co-labeling of the biocytin-filled neurons with the neurochemical markers expected on the basis of the physiological recordings. Thus, we are confident that the restaining procedure does not lead to loss of specificity in labeling.
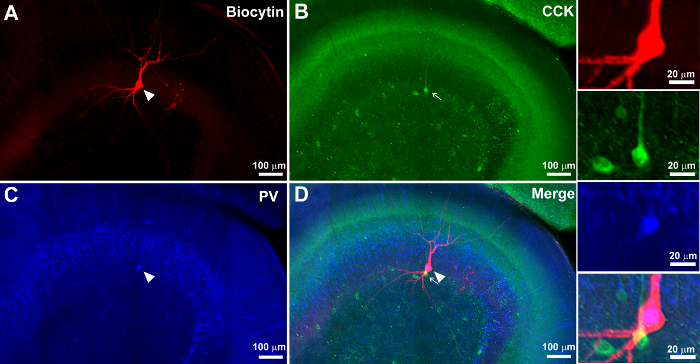
Figure 4. Lack of Overlap between Mutually Exclusive Markers on the Second Immunostaining Following Prior Immunostaining and Mounting of Sections. (A – C) Confocal images at 10X showing a biocytin-filled neuron (A) in red, indicated by an arrowhead, and CCK immunoreactivity in green (B), indicated by the arrow. Note the lack of overlap. The same section was processed for parvalbumin (PV) immunostaining in blue after more than 2 months (C). The overlay of images is shown in D. Note that the PV axons are distributed in the granule cell layer, with CCK in the molecular layer in the expected nonoverlapping pattern. Also note the lack of colocalization of the two markers in neuronal somata. The zoomed images to the right illustrate that the biocytin-labeled neuron expresses PV (arrowhead) and not CCK (arrow). Scale bar: 100 µm (A – D), 20 µm (panels to the right). Please click here to view a larger version of this figure.
One common reason for sub-optimal biocytin fills is the accidental severing of the neuronal processes with the recording pipette while approaching the cell. A lost appendage of the cell appears as a bleb of biocytin at the end of an axon, as illustrated in Figure 1 (green arrow). Axonal blebs are often visualized when patching superficial cells, whose axons may be severed during slicing procedures. However, optimization of the plane of slicing and knowledge of the axonal distribution pattern of the targeted cell class can aid in avoiding approaching the soma from the direction of the putative axon, which can also sever the axon, resulting in the appearance of an axonal bleb. In most cell types, it is preferable to approach the cell body along the Z-axis (white arrow with mark). A second issue concerns the application of excessive positive pressure to the pipette while approaching the cell. This can not only damage the cell and its connections but also causes the internal solution containing biocytin to spill around the cell, leading to high background fluorescence. Background staining also increases when the time taken to approach the cell is prolonged or when slice quality is not optimal, which can compromise the ability to define cellular morphology (Figure 5A). It is possible to pull the soma out while dislodging the pipette. This will result in a lack of somatic morphology (Figure 5B). To avoid pulling out the cell, move the pipette up and out in small steps using the "fine" movement settings of the manipulator. If the cell appears to be attached to the pipette through a long membrane extension, tap/flick the manipulator gently to dislodge the pipette from the cell. Avoid using the coarse manipulator settings until the cell has fully dislodged.
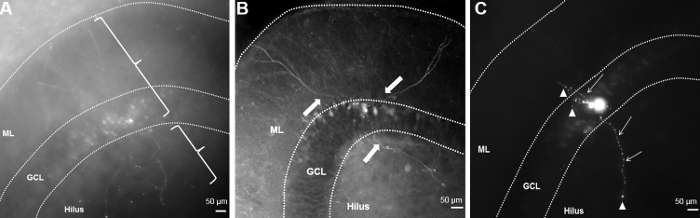
Figure 5. Images Illustrate Unsuccessful Biocytin Fills. (A) Image showing high background fluorescence due to excessive positive pressure in the recording pipette, resulting in poor cell visibility. Finer structures of neuronal processes, including axons, are visualized beyond the region of the spill. Also, notice that soma has been pulled out. (B) A neuron with the soma pulled out during pipette detachment. Arrows show successful fills of the axon and dendrites, whereas the soma and appendages proximal to the arrows have not been recovered. (C) A cell patched in a superficial layer of the slice (<50 µm from surface) results in filled cells exhibiting cut axon and dendrites (arrowheads). Notice the beading (arrows) of granule cell dendrites, indicating poor cell health (C). Scale bar: 50 µm. Please click here to view a larger version of this figure.
Finally, in order to recover complete cellular morphology without severed axons or dendrites, it is best to target cell >50 µm deep to the slice surface. Superficial cells may have processes that have been cut and may lack the normal complement of synaptic inputs.
The most common pitfalls in the restaining procedures are associated with the removal of the coverslip and the recovery of the section without damaging the tissue. A second issue concerns antibody penetration into thick tissue, especially when incubated overnight. In several instances, better penetration has been achieved by incubating in the primary antibody for up to 72 h at 4 °C on an orbital shaker placed in a cold room. Although re-sectioning the tissue is the most effective procedure to ensure antibody penetration, prolonging the duration of incubation in primary antibody or increasing the detergent concentration to 0.5 – 1% Triton-X100 may also aid antibody penetration24. Since most recorded neurons are located within 50 – 100 µm from the surface of the section, we find that the staining of thick sections is adequate to label biocytin-filled somata or processes at this level. However, it is recommended that the extent of antibody labeling is checked on each section before interpreting negative co-localization results.
Discussion
Critical Steps within the Protocol
Filling the patched cell with biocytin is the most crucial step to ensure the full recovery of the morphology. For full recovery of the cell, it is essential to select an optimal slice orientation to minimize the severing of processes during slicing. This orientation may differ based on the circuit and cell type under examination. Next, it is essential to allow adequate time for the biocytin to diffuse to the dendrites and axons. Some dyes, such as neurobiotin, can also penetrate neurons coupled to the recorded cell through gap junctions. The size of the pipette tip also needs to be optimized, as the soma tends to be pulled out when the opening is large and biocytin loading is compromised when the pipette tip is too small. Additionally, appropriate procedures while disengaging from whole-cell mode are essential to recover the soma. After recording for 24 h, the subsequent incubation of the slices in 4% PFA followed by PBS ensures that the cross-linking of the tissues is reduced, which is critical for immunohistochemistry. Storage of the slices at 4 °C is also crucial for maintaining slice integrity. However, prolonged storage in PFA reduces eventual success with antibody-labeling procedures. Optimizing the concentration and duration of the primary antibody incubation is important, as these will differ for individual antibodies, and some antibodies may require from 3 – 5 d of incubation for optimal penetration in thick tissue. To ensure that the slices are undamaged before the secondary primary antibody staining, adequate care is needed during the removal of the cover slip.
Modifications and Troubleshooting
We have observed that there is no set duration between the first and second stain. Sections have been re-stained months later on the same tissue and with more than one stain. In our recent works, we compared sections processed using the re-staining method with those using standard immunostaining protocols and find no obvious difference in the staining patterns or the robustness of biocytin fills6,9,25,26.
While it remains to be tested, it is possible that re-staining may be performed more than once, with tissue integrity being a limiting factor for repeated handling. The availability of discrete fluorophores also poses a limit for the number of antigens that can be visualized. Therefore, careful handling of the tissue is advised. An additional process that remains to be tested is restaining using antigen recovery protocols (for surface receptors). Since the integrity of the tissue is retained after removal from the cover slip, we expect that the procedure should also work in staining protocols that recommend antigen-recovery. In this regard, physiological recordings often yield cells with no known neurochemical markers6,9. The procedures detailed here allow for the recovery of their morphology based on biocytin immunostaining. While the neurochemical identity may not be known a priori, re-staining for potential markers could facilitate identification of markers for neurons that have been described on the basis of morphology and electrical properties.
Limitations of the Technique
A limitation associated with performing immunohistochemical staining on thick sections is the insufficient penetration of the antibody through the complete thickness of the section, especially with standard overnight incubation in the primary antibody. Moreover, different antibodies and streptavidin, used to reveal biocytin, can have differential tissue penetration. Thus, it is recommended that trial runs are performed to assess the depth of antibody penetration and to modify the incubation times, temperatures, and antibody and Triton X-100 concentrations in order to achieve optimal slice penetration and staining for each antibody. It should also be noted that successful labeling with one antibody or with streptavidin-biocytin does not imply that a second antibody will penetrate to the same depth of the section. Therefore, care must be taken to verify the penetration of each antibody to the level at which the biocytin-labeled cell processes are examined for co-localization with the neurochemical marker. If the penetration is inadequate, the slice can be re-sectioned at <60 µm for immunostaining. Note, however, that since the slices were already processed for biocytin, the morphology of the cell can be captured by confocal imaging to eliminate the possible loss of anatomical data during re-sectioning and to facilitate neuronal reconstruction. While it is possible that the age and animal species may alter the degree of antibody penetration, we have successfully used these procedures in rat tissue from 30-day- and over 60-day-old rats and in mice.
A second limitation is that biocytin is not inherently fluorescent and cannot be used for live imaging and real-time morphological characterization during recording. Alternative unconjugated fluorophores and Lucifer yellow have been utilized for live imaging of the cells27. They are not permanent and lose fluorescence upon fixation, making it impractical for post-hoc neurochemical identification of the recorded neuron. Recently, fluorophores with biocytin have been introduced to overcome this limitation and can be used for live imaging as well as for post-hoc processing in double- or triple-labeling, as detailed here. Interestingly, to date, biocytin fills have resulted in better electrical and staining properties for combined electrophysiological and anatomical studies when compared to Lucifer yellow28.
Significance of the Technique with Respect to Existing/Alternative Methods
The major advantages of using the protocol detailed here are that, unlike resectioning, the biocytin-filled cell structure and processes remain intact and do not lead to the permanent loss of tissue, nor to a need for laborious reconstructions from multiple sections. Thus, a single-cell fill followed by immunocytochemistry can help in the morphological reconstruction and identification of specific neurochemical markers.
Future Applications or Directions after Mastering This Technique
In all, we present a novel practical approach for the recovery of morphology and post-hoc double- and triple-immunolabeling of neurons following electrophysiological recordings. The process is particularly advantageous for finding new markers as knowledge evolves, re-evaluating neurons that were previously filled, visualized, and imaged. It is particularly advantageous because the immunohistochemistry can be performed weeks or even months later, saving tissue and antibodies. The method does not require any special chemicals and can safely be performed in any laboratory.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors would like to acknowledge the support from NIH/NINDS R01 NS069861 and NJCBIR CBIR14IRG024 to VS.
Materials
| NaCl | Sigma | S7653 | Immunostaining |
| KCL | Fluka | 60129 | Immunostaining |
| Na2HPO4 | Sigma | S7907 | Immunostaining |
| KH2PO4 | Sigma | 229806 | Immunostaining |
| Triton X-100 | Sigma | T8787 | Immunostaining |
| Guinea pig anti CB1 | Sigma | Af530-1 | Immunostaining |
| Mouse anti CCK | CURE, UCLA | courtesy of G. Ohning | Immunostaining |
| Rabbit anti Parvalbumin | Swant | PV27 | Immunostaining |
| Streptavidin, Alexa Fluor conjugate | Molecular Probes | S11227 | Immunostaining |
| Normal goat serum | Sigma | G9023 | Immunostaining |
| Vectashield | Vector Labs | H-1000 | Immunostaining |
| Secondary Antibodies | Invitogen Molecular probes | Alexa Fluor conjugated dyes | Immunostaining |
| Labnet orbit low speed shaker | Bioexpress | S-2030-LS | Immunostaining |
| Forceps | Dumont | 11231-30 | Immunostaining |
| Slide folders | EMS | 71520 | Immunostaining |
| Vibratome VT 1200 S | Leica | 14048142066 | Electrophysiology |
| Multiclamp 700B amplifier | Molecular devices | Multiclamp 700B | Electrophysiology |
| pCLAMP 10 Software | Molecular devices | pCLAMP 10 | Electrophysiology |
| Digitizer | Molecular Devices | Digidata 1440 digitizer | Electrophysiology |
| Filter tips | Nalgene | 171-0020 | Electrophysiology |
| Sonicator | Fisher Scientific | 15-335-100 | Electrophysiology |
| Microloaders | Eppendorf | 930001007 | Electrophysiology |
| Biocytin | Sigma | B4261 | Electrophysiology |
References
- Garcia-Lopez, P., Garcia-Marin, V., Freire, M. The histological slides and drawings of cajal. Front Neuroanat. 4, 9 (2010).
- Klausberger, T., Somogyi, P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 321 (5885), 53-57 (2008).
- Petilla Interneuron Nomenclature, G., et al. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci. 9 (7), 557-568 (2008).
- Armstrong, C., Soltesz, I. Basket cell dichotomy in microcircuit function. J Physiol. 590 (4), 683-694 (2012).
- Povysheva, N. V., Zaitsev, A. V., Gonzalez-Burgos, G., Lewis, D. A. Electrophysiological heterogeneity of fast-spiking interneurons: chandelier versus basket cells. PLoS One. 8 (8), e70553 (2013).
- Gupta, A., Elgammal, F. S., Proddutur, A., Shah, S., Santhakumar, V. Decrease in tonic inhibition contributes to increase in dentate semilunar granule cell excitability after brain injury. J Neurosci. 32 (7), 2523-2537 (2012).
- Fish, K. N., Hoftman, G. D., Sheikh, W., Kitchens, M., Lewis, D. A. Parvalbumin-containing chandelier and basket cell boutons have distinctive modes of maturation in monkey prefrontal cortex. J Neurosci. 33 (19), 8352-8358 (2013).
- Yu, J., Swietek, B., Proddutur, A., Santhakumar, V. Dentate cannabinoid-sensitive interneurons undergo unique and selective strengthening of mutual synaptic inhibition in experimental epilepsy. Neurobiol Dis. 89, 23-35 (2016).
- Yu, J., Swietek, B., Proddutur, A., Santhakumar, V. Dentate total molecular layer interneurons mediate cannabinoid-sensitive inhibition. Hippocampus. 25 (8), 884-889 (2015).
- Varga, C., et al. Functional fission of parvalbumin interneuron classes during fast network events. Elife. 3, (2014).
- Kawashima, T., Okuno, H., Bito, H. A new era for functional labeling of neurons: activity-dependent promoters have come of age. Neural Circuits Revealed. , 109 (2015).
- Krook-Magnuson, E., Luu, L., Lee, S. H., Varga, C., Soltesz, I. Ivy and neurogliaform interneurons are a major target of mu-opioid receptor modulation. J Neurosci. 31 (42), 14861-14870 (2011).
- Szabadics, J., Soltesz, I. Functional specificity of mossy fiber innervation of GABAergic cells in the hippocampus. J Neurosci. 29 (13), 4239-4251 (2009).
- Iball, J., Ali, A. B. Endocannabinoid Release Modulates Electrical Coupling between CCK Cells Connected via Chemical and Electrical Synapses in CA1. Front Neural Circuits. 5, 17 (2011).
- Chen, X., Cho, D. -. B., Yang, P. -. C. Double staining immunohistochemistry. N Am J Med Sci. 2 (5), 241-245 (2010).
- Ranjan, A. K., et al. Cellular detection of multiple antigens at single cell resolution using antibodies generated from the same species. Journal of immunological. 379 (1), 42-47 (2012).
- Fuccillo, D. A., Sever, J. L. . Concepts in Viral Pathogenesis II. , 324-330 (1986).
- Buchwalow, I. B., Minin, E. A., Boecker, W. A multicolor fluorescence immunostaining technique for simultaneous antigen targeting. Acta Histochem. 107 (2), 143-148 (2005).
- Qi, G., Radnikow, G., Feldmeyer, D. Electrophysiological and morphological characterization of neuronal microcircuits in acute brain slices using paired patch-clamp recordings. JoVE (Journal of Visualized Experiments. (95), e52358 (2015).
- Walz, W. . Patch-clamp analysis : advanced techniques. 2nd edn. , (2007).
- Molnar, P. . Patch-clamp methods and protocols. 403, (2007).
- Martina, M., Taverna, S. . Patch-clamp methods and protocols. , (2014).
- Booker, S. A., Song, J., Vida, I. Whole-cell patch-clamp recordings from morphologically- and neurochemically-identified hippocampal interneurons). J Vis Exp. (91), e51706 (2014).
- Jinno, S., Kosaka, T. Immunocytochemical characterization of hippocamposeptal projecting GABAergic nonprincipal neurons in the mouse brain: a retrograde labeling study. Brain research. 945 (2), 219-231 (2002).
- Yu, J., Proddutur, A., Swietek, B., Elgammal, F. S., Santhakumar, V. Functional Reduction in Cannabinoid-Sensitive Heterotypic Inhibition of Dentate Basket Cells in Epilepsy: Impact on Network Rhythms. Cereb Cortex. , (2015).
- Proddutur, A., Yu, J., Elgammal, F. S., Santhakumar, V. Seizure-induced alterations in fast-spiking basket cell GABA currents modulate frequency and coherence of gamma oscillation in network simulations. Chaos. 23 (4), 046109 (2013).
- Scharfman, H. E. Differentiation of rat dentate neurons by morphology and electrophysiology in hippocampal slices: granule cells, spiny hilar cells and aspiny ‘fast-spiking’ cells. Epilepsy Res Suppl. 7, 93-109 (1992).
- Tasker, J. G., Hoffman, N. W., Dudek, F. E. Comparison of three intracellular markers for combined electrophysiological, morphological and immunohistochemical analyses. J Neurosci Methods. 38 (2-3), 129-143 (1991).

