An In Vitro Caseum Binding Assay that Predicts Drug Penetration in Tuberculosis Lesions
Summary
Here we describe a rapid equilibrium dialysis (RED) method to measure drug binding to caseum from pulmonary tuberculosis lesions and cavities. The protocol is also used with a foamy macrophage-derived matrix that is an effective surrogate to caseum.
Abstract
The eradication of tuberculosis disease requires drug regimens that can penetrate the multiple layers of complex pulmonary lesions. Drug distribution in the caseous cores of cavities and lesions is especially crucial because they harbor subpopulations of drug-tolerant bacteria also commonly referred to as persisters. Existing methods for the measurement of drug penetration in tuberculosis lesions involve costly and time-consuming in vivo pharmacokinetic studies coupled to bioanalytical or imaging techniques. The in vitro measurement of drug binding to caseum macromolecules was proposed as an alternative to such techniques since this binding hinders the passive diffusion of drug molecules through caseum. Rapid equilibrium dialysis is a fast and reliable system for performing plasma protein and tissue binding studies. In this protocol, we used a rapid equilibrium dialysis (RED) device to measure drug binding to homogenates of caseum that is excised from the lesions and cavities of tuberculosis-infected rabbits. The protocol also describes how to generate a surrogate matrix from lipid loaded THP-1 macrophages to use in place of caseum. This caseum/surrogate binding assay is an important tool in tuberculosis drug discovery and can be adapted to help study drug distribution in lesions or abscesses caused by other diseases.
Introduction
The treatment of pulmonary tuberculosis disease requires effective distribution of drugs into different types of lesions. Necrotic lesions and cavities contain caseous centers that harbor subpopulations of drug-tolerant or 'persistent' bacteria.1,2 The cavitary disease is associated with inferior cure rates and poor prognosis.3,4 Previous studies have shown, using quantitative and imaging techniques, that the ability to penetrate caseum varies significantly from one drug class to another.5,6 These methods, however, require the use of animal infection models that are slow and tedious. An in vitro assay that measures drug binding to ex vivo caseum was designed. This binding was found to inversely correlate with drug penetration in caseous granulomas and, hence, is used as a predictive tool.7
Equilibrium dialysis is regarded as the gold standard approach to plasma protein binding studies. The rapid equilibrium dialysis (RED) device provides a quick, easy-to-use and reliable system for performing such assays.8 The device is made up of two components: single-use, disposable inserts consisting of 2 chambers separated by a vertical cylinder of semi-permeable membrane; and reusable base plates that can hold up to 48 inserts at a time. The dialysis membrane has an 8 kDa molecular weight cut-off (MWCO) that is ideal for drug-macromolecule binding studies. The high surface area-to-volume ratio of the membrane compartment allows rapid dialysis and equilibration. Both the inserts and the base plate have been validated for minimal non-specific binding. The combination of the RED device with bioanalytical techniques provides accurate estimations of the unbound fractions of drugs in plasma.8,9
Although originally designed to measure plasma protein binding, the RED device has been used in several tissue binding studies using homogenates.10,11 In this protocol, we measure drug binding to caseum, the necrotic debris excised from the necrotic lesions and cavities of tuberculosis-infected rabbits. The acellular and non-vascular nature of caseous material makes it easy to homogenize into a homogenous suspension that is compatible with the assay.
Given that caseum is tedious to produce and hard to come by, the protocol has also been validated for use with a surrogate matrix that is prepared from foamy macrophages. THP-1 monocyte-derived macrophages are induced with oleic acid to accumulate multiple lipid bodies that give them their 'foamy' appearance. These lipid-loaded cells are harvested and processed to produce a matrix that we use as a surrogate to caseum. This study has shown that drug binding to this surrogate matrix correlates well with binding to caseum, effectively mimicking the in vivo process that hinders drug penetration in the caseous core of granulomas and cavities.
Protocol
All animal studies were carried out in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health with approval from the Institutional Animal Care and Use Committee of the NIAID (NIH), Bethesda, MD. All studies involving M. tuberculosis were performed in a laboratory with biosafety containment level 3 (BSL-3).
1. Rabbit Infection Model and Caseum Collection
- Infect New Zealand white rabbits with M. tuberculosis using a nose-only aerosol exposure system as previously described.12,13 Allow the infection to progress for 12-16 weeks. Sedate the rabbits with 35 mg/kg ketamine and 5 mg/kg xylazine intramuscularly, euthanize the rabbits with 0.22 mL/kg pentobarbital sodium and phenytoin sodium intravenously and proceed with the necropsies.
- Using tweezers and a scalpel, remove lungs from the chest cavity. From each lung lobe, dissect out individual cavities and large necrotic granulomas using a scalpel. Carefully scrape off caseum from the cavity and granuloma walls. Weigh, record and store samples in 2 mL screw-capped tubes at -20 °C until ready for use.
- Gamma-irradiate the infectious caseum samples at 3 MegaRad on dry ice to render them uninfectious and safe for use in a BSL-2 lab.
2. In Vitro Generation of Caseum Surrogate from THP-1 Cells
- Grow THP-1 monocytes in RPMI 1640 medium (2 mM L-glutamine and 10% fetal bovine serum) in T175 cell culture flasks (80 mL/flask). Incubate the flasks in a 5% CO2 atmosphere at 37 °C for 3-4 days.
- Centrifuge the culture from a T175 flask in two 50 mL conical tubes at 150 x g for 5 min. Discard the supernatant and suspend the pellet in 10 mL of RPMI 1640 media.
- Pipette 5 µL of this culture into a 1.5 mL tube containing 45 µL of trypan blue. Mix thoroughly by pipetting. Transfer 10 µL to a hemocytometer and count the number of viable THP-1 monocytes (unstained) using a light microscope (10X magnification). Calculate the number of viable cells per mL of the culture. Dilute it with RPMI media to the final density of 1.25 x 106 cells/mL.
- Load 40 mL of the culture on a large cell culture plate (50 x 106 cells/plate). Add 40 µL of 100 µM PMA (phorbol 12-myristate13-acetate prepared in ethanol) and allow cells to adhere overnight in the incubator.
NOTE: Final concentration of PMA is 100 nM. - Dilute pure oleic acid (OA) (0.89 g/mL) in ethanol to the concentration of 0.1 M (i.e. 31.7 µL OA in 968.3 µL ethanol). Dilute this solution in fresh pre-warmed RMPI media to a concentration of 10 mM. Dilute this OA suspension to 0.4 mM (final working concentration) in RPMI medium pre-warmed to 37 °C.
- Remove the existing media and non-adhered cells from the cell culture plates and gently add 40 mL of 0.4 mM OA to the THP-1 macrophages (THP-M). Incubate at 37 °C in the incubator overnight.
- Use a light microscope at 40x magnification to visually confirm the presence of numerous lipid body inclusions in each THP-M. Remove all RPMI medium from the cell culture plates and gently wash the adherent cells twice with phosphate buffered saline (PBS) using a 50 mL serological pipette.
NOTE: Lipid bodies appear as small, clear, spherical structures in the cytoplasm of the THP-M. - Add 40 mL of 5 mM ethylenediaminetetraacetic acid (EDTA) in PBS to each plate. Incubate for 15 min at 37 °C.
- Detach the foamy macrophages (FM) by repeatedly pipetting up and down over the surface of the whole plate using a 10-mL serological pipette. Transfer the cell suspension to a 50-mL conical tube and spin down at 150 x g for 5 min.
- Resuspend the cell pellet in 10 mL of PBS (third PBS wash) and transfer to a pre-weighed 15-mL conical tubes. Spin down again at 150 x g for 5 min. Carefully aspirate the supernatant using a serological pipette and discard.
- Subject the FM pellets to 3 freeze-thaw cycles to lyse the cells and incubate them at 75 °C for 20-30 min to denature proteins in the matrix. Store the pellets at -20 °C until ready for use.
3. Rapid Equilibration Dialysis (RED) Assay
- Prepare 10 mM stock solutions of all test compounds in dimethyl sulfoxide (DMSO). Dilute to 500 µM working solutions in DMSO prior to each assay.
- Weigh the tube containing the caseum surrogate pellet. Subtract the weight of the empty tube to derive the weight of the pellet alone. Add 2-3 metal beads per tube and, using a tissue homogenizer at 1,200 strokes/min for 1 min, disrupt caseum or the surrogate matrix in PBS (1:9 w/v) to achieve 10x diluted suspension of each matrix.
- Spike 6.5 µL of the 500 µM solution of the test compound into 643.5 µL of the homogenate to achieve the final concentration of 5 µM (≤1% DMSO) and vortex.
- Place the RED inserts into the base plate. Add 200 µL of the drug-spiked matrix into the donor chamber (red ring) of each RED insert and 350 µL of PBS into each receiver chamber. Prepare 3 inserts for each test compound (triplicate samples). Seal plate with an adhesive plate seal and incubate at 37 °C on the thermomixer at 200 rpm (1 x g) for 4 h.
- After the incubation, gently mix the contents of the donor and receiver chambers by pipetting up and down 2-3 times. Pipette out 20 µL aliquots of homogenate from the donor chambers and add to 20 µL of clean PBS in a 1.5 mL tube (1:1). Similarly, pipette out 20 µL aliquots of PBS samples from the receiver chambers and add to 20 µL of clean homogenate (matrix matching).8
NOTE: Matrix-matching eliminates the need for 2 separate calibration curves (in homogenate and PBS) to be made for quantitative analysis. Contents of the donor chamber may sediment over time. Gently mix the contents by pipetting prior to removing aliquots.
4. LC-MS Quantification and Data Analysis
- Add 160 µL of 1:1 methanol:acetonitrile containing 500 ng/mL diclofenac or 10 ng/mL verapamil (internal standard) to each tube and vortex to precipitate proteins. Centrifuge at 10,000 x g for 5 min to sediment the precipitate and transfer supernatants into 96-well deep-well plates for liquid chromatography-mass spectrometry (LCMS) analysis.7
- Build calibration curves from 1-1,000 nM for each test compound while keeping the same matrix composition as the samples above. Quantify the concentration of test compound in samples from the donor and receiver chambers using an LC-MS method.
- Calculate the unbound fraction (fu) of the drug in diluted matrix using equation 1. Calculate the fu in undiluted matrix using equation 2 (D = dilution factor of 10).14
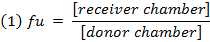
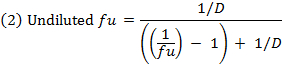
- Check the recovery (mass balance) of each compound using equation 3 to identify compounds with stability/metabolism/non-specific binding issues.
NOTE: Recovery typically falls between 70% and 130%.15
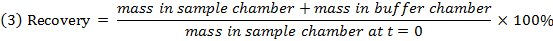
Representative Results
Using this protocol, we have tested hundreds of tuberculosis drug development compounds for their predicted efficiency at penetrating caseum. Figure 1 visualizes the basic concepts of the RED assay. The dialysis membrane of the RED inserts allows for unbound small molecules to diffuse from the donor well to the receiver well, finally achieving an equilibrium between both compartments. Small molecules that are bound to macromolecules such as proteins or lipids are trapped in the donor well. Quantitative analysis of the samples from both compartments allows for the calculation of the unbound fraction (fu) of each small molecule in the caseum or surrogate matrix. This fraction has been shown to directly correlate with drug penetration in caseum in vivo.7 As Figure 2 illustrates using MALDI (Matrix-assisted laser desorption/ionization) mass spectrometry images, the higher the fu, the more extensively the drug diffuses into caseum. Hence, this agrees with the hypothesis that as drug molecules diffuse into the outer rim of caseum from its interface with cellular layers of the lesion, binding to caseous macromolecules sequesters the drug, preventing it from further diffusing into the caseous core. A sample panel of 18 anti-tuberculosis drugs is listed in Table 1 along with their fu in both caseum and the surrogate. As Figure 3 illustrates, the matrix prepared from THP-1-derived foamy macrophages is an effective surrogate to true caseum. Binding in both matrices strongly correlates with each other.
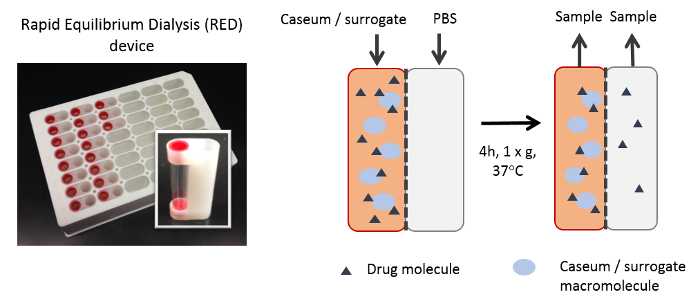
Figure 1: Illustration of the RED assay. During incubation, drug molecules that remain unbound to macromolecules in the matrix diffuse across the dialysis membrane. Over time, an equilibrium is established between the concentration of unbound drug molecules in both the donor and receiver wells. All macromolecules (proteins, lipids, etc.) and bound drug molecules are trapped within the donor chamber. Please click here to view a larger version of this figure.
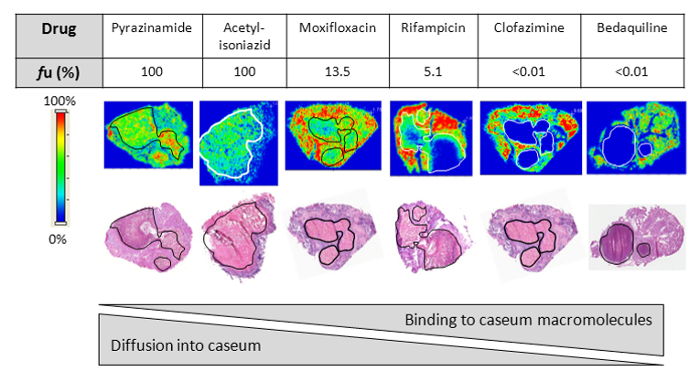
Figure 2: Drug penetration in vivo. Relationship between the fraction unbound (fu) and diffusion into caseum in vivo as determined by MALDI (matrix-assisted laser desorption/ionization) mass spectrometry imaging for six anti-tuberculosis drugs. Ion maps are of representative lung lesions and the signal intensity is indicated by the scale bar to the left. Hematoxylin and Eosin (H&E) staining of adjacent sections is shown below the ion maps. Black/white contour lines highlight the caseous center of each lesion. Reprinted (adapted) with permission from Sarathy et al.7 Please click here to view a larger version of this figure.
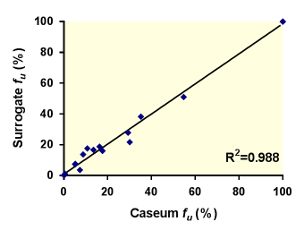
Figure 3: Correlation between the unbound fractions (fu) of 18 anti-tuberculosis drugs in caseum and the surrogate matrix. The best-fit line was determined using linear regression. The goodness of fit is expressed as the R2 value. Reprinted (adapted) with permission from Sarathy et al.7 Please click here to view a larger version of this figure.
| Caseum fu (%) | Surrogate fu (%) | |
| Ethambutol | 35.2 ± 4.4 | 38.8 ± 5.6 |
| Isoniazid | >99.9 | >99.9 |
| Acetyl-isoniazid | >99.9 | >99.9 |
| p-aminosalicylic acid | 54.7 ± 1.4 | 51.1 ± 11.2 |
| Rifampicin | 5.13 ± 0.2 | 7.3 ± 0.6 |
| Rifapentine | 0.5 ± 0.1 | 1.1 ± 0.1 |
| Moxifloxacin | 13.5 ± 3.7 | 16.8 ± 1.8 |
| Levofloxacin | 8.34 ± 0.4 | 16.3 ± 3.1 |
| Gatifloxacin | 16.3 ± 4.2 | 18.8 ± 2.9 |
| Linezolid | 29.3 ± 3.6 | 27.9 ± 2.2 |
| Posizolid | 10.7 ± 1.7 | 17.6 ± 4.5 |
| Sutezolid | 30.1 ± 8.7 | 21.7 ± 1.7 |
| Radezolid | 5.2 ± 0.8 | 7.6 ± 1.9 |
| Tedizolid | 8.8 ± 1.8 | 13.7 ± 2.7 |
| Clofazimine | <0.01 | <0.01 |
| Bedaquiline | <0.01 | <0.01 |
| PA-824 | 7.31 ± 2.2 | 3.6 ± 0.2 |
| OPC67683 | 0.04 ± 0.02 | 0.02 ± 0.002 |
Table 1: Fraction unbound (fu) of the 18 TB drugs tested in the caseum and surrogate binding assay. All results are expressed as the mean ± standard deviation. Reprinted (adapted) with permission from Sarathy et al.7
Discussion
Pulmonary necrotic lesions and cavities in tuberculosis-infected patients contain subpopulations of bacteria that are recalcitrant to drug treatment. The caseous cores of these structures are particularly responsible for harboring these persisters in an extracellular environment.16 Favorable distribution of anti-bacterial agents into these remote locations is believed to be an important determinant of tuberculosis drug efficacy. Prior to the validation of this protocol, there were no available in vitro techniques that could predict the distribution of drug discovery compounds in caseous lesions. As a result, the evaluation of drug distribution in these critical foci was heavily dependent on animal infection models, lesion-centric pharmacokinetic studies and MALDI mass spectrometry imaging techniques.5,17
The in vitro assay described here is a fast and effective way to predict drug penetration in tuberculosis lesions without the tedium and cost of in vivo pharmacokinetic studies. The protocol is a modification of the plasma protein binding assay which was originally the intended use of the RED device. Homogenization of tissue samples to a pipette-able consistency allows for tissue protein binding to be measured using the same device. This protocol can be conducted with caseum if it is available from animal infection models. As Figure 1 explains, drug molecules bind to macromolecules in caseum/surrogate homogenate, keeping it in the donor chamber, whereas the unbound drug molecules are free to diffuse across the semi-permeable membrane into the receiver chamber. The unbound fraction (fu) equilibrates between both compartments during the incubation period. As Figure 2 illustrates, very highly bound compounds (fu ≤ 1%) that are almost entirely trapped within the donor chamber are not effective at permeating caseum. On the other hand, less-bound compounds (1% < fu < 100%) are able to diffuse into the caseous core to varying degrees; the higher the fu, the more extensively it permeates the necrotic area.
The protocol can also be used with a surrogate matrix derived from lipid-loaded THP-1 macrophages that have been induced with oleic acid. Several conditions for the induction of lipid body accumulation were tested on THP-1s. These include hypoxia, exposure to irradiated M. tuberculosis and exposure to trehalose 6,6'-dimycolate (TMD) derived from the mycobacterial cell wall. The final oleic acid incubation concentration of 400 µM was proven to induce peak lipid body formation without compromising cell viability and adhesion.7 The dilution of oleic acid in pre-warmed media is critical to ensure maximal dissolution of the fatty acid. The resulting surrogate matrix has a comparable cholesterol content to true caseum, but higher and lower triglyceride and protein contents, respectively.7 As Figure 3 illustrates, the binding of 18 anti-tuberculosis compounds to the surrogate matrix strongly correlates with their binding to caseum. The recent study that also included non-commercial compounds with undefined bactericidal activity proved that the FM-derived matrix is an effective surrogate to caseum.7 By removing the need for animal infection models, the surrogate matrix successfully increases the throughput of this assay while making it more cost-effective.
This versatile protocol can also be used to test multiple compounds simultaneously (cassette testing). We have previously shown that testing 5 drug-combinations, all at 5 µM, in each insert using this protocol can effectively rank compounds based on their caseum binding affinities while saving time and materials. A sixth control compound such as moxifloxacin may be added to the cassette as an internal control to ensure consistency between assays and batches of caseum/surrogate.7 Importantly, the protocol can be run with as little as 20 µL of a 10 mM compound stock (<<1 mg of a compound). This is a huge advantage early on in the drug discovery process when chemists synthesize limited amounts of each compound for screening purposes. The RED device apparatus are automation friendly; the microplate footprint of the base plate makes it compatible with automated systems designed to handle standard 96-well plates.
Although the surrogate matrix has been shown to successfully mimic the binding properties of ex vivo caseum (Figure 3) we acknowledge that it may inaccurately profile certain compounds. THP-1 macrophages are commonly used in in vitro techniques to simulate alveolar macrophages in lung tissue. However, the surrogate matrix derived from THP-1 does not have the same composition as the necrotic caseous material in lung lesions. As a result, certain classes of compounds may interact differently with the surrogate matrix. Moreover, the success of the assay is contingent on the stability of the test compound in the caseous matrix. Caseum is a biological matrix that shares many components with plasma, and hence this protocol is susceptible to compound degradation issues in the same way many plasma-based assays are as well. In this respect, the THP-M-derived surrogate matrix is preferable because the denaturation step in the protocol removes enzyme activity, improving the total recovery at the end of the assay.
The caseum surrogate binding assay is a useful tool in tuberculosis drug discovery and is being used in multiple lead optimization programs. It can also help design more effective combination regimens based on the ability of individual drugs to partition into the various layers of multiple lesion-types. The protocol could also be adapted to facilitate drug discovery for other diseases that are characterized by the formation of lesions or abscesses where drug penetration is limited but critical for successful treatment.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We wish to thank Johnson & Johnson, the TB Alliance, Astra Zeneca, Rib-X and Trius Therapeutics for providing bedaquiline, PA-824 (pretomanid), AZD5847, radezolid and tedizolid, respectively. Brendan Prideaux, Matthew Zimmerman, Stephen Juzwin, Emma Rey-Jurado, Nancy Ruel, Leyan Li and Danielle Weiner provided support with MALDI analysis, bioanalytical methods, preparation of the caseum surrogate, chemical synthesis, and isolation of rabbit caseum. This work was conducted with funding from the Bill and Melinda Gates Foundation, award # OPP1044966 and OPP1024050 to V. Dartois, NIH Shared Instrumentation Grant S10OD018072, as well as joint funding from the Bill and Melinda Gates Foundation and Wellcome Trust for A Centre of Excellence for Lead Optimization for Diseases of the Developing World to P. Wyatt.
Materials
| New Zealand White Rabbits | Covance | – | |
| HN878 Mycobacterium tuberculosis | BEI Resources | NR-13647 | |
| Ketathesia (Ketamine) 100mg/mL C3N | Henry Schein Animal Health | 56344 | |
| Anased (Xylazine) 100mg/mL | Henry Schein Animal Health | 33198 | |
| Euthasol (pentobarbital sodium and phenytoin sodium) Solution | Virbac | 710101 | |
| THP-1 monocytic cell line | ATCC | ATCC TIB-202 | |
| 175cm² TC-Treated Flask (T175) | Fisher Scientific | T-3400-175 | |
| RPMI 1640 media w/o glutamine | Fisher Scientific | MT-15-040-CV | |
| Hyclone Fetal Bovine Serum, Gamma irradiated | Fisher Scientific | SH3091003IR | |
| Hyclone L-glutamine, 200mM | Fisher Scientific | SH3003401 | |
| Cellstar TC dish, 145x20mm, vented | Fisher Scientific | T-2881-1 | |
| Phorbol 12-myristate13-acetate (PMA) | Fisher Scientific | BP685-1 | |
| Ethylenediaminetetraacetic acid | Sigma | E6758 | |
| Oleic acid | Fisher Scientific | ICN15178101 | |
| Pierce RED Device Reusable Base Plate | Fisher Scientific | PI-89811 | |
| Pierce RED Device Inserts, 50/box | Fisher Scientific | PI-89809 | |
| Pierce RED insert removal tool | Fisher Scientific | 89812 | |
| Adhesive plate seal | Fisher Scientific | 08-408-240 | |
| PBS, pH7.4, 10 X 500 mL (Gibco) | Life Technologies | 10010-049 | |
| DMSO | Sigma | 472301 | |
| Acetonitrile | Sigma | 34998 | |
| Methanol | Sigma | 34860 | |
| Verapamil hydrochloride | Sigma | V4629 | |
| Diclofenac sodium salt | Sigma | 93484 | |
| Trypan Blue Solution, 0.4% | Fisher Scientific | 15-250-061 | |
| Ethanol, 200 proof | Fisher Scientific | 04-355-451 | |
| 2010 Geno/Grinder | SPEX SamplePrep | 2010 | |
| Bead Mill Homogenizer Accessory, Metal Bulk Beads | Fisher Scientific | 15-340-158 | |
| 484R Cobalt 60 Irradiator | JL Shepard | 7810-484-1 | |
| INCYTO C-Chip Disposable Hemacytometers | Fisher Scientific | 22-600-100 | |
| Upright Light Microscope | Leica | DM1000 | |
| Binary Liquid Chromatography system | Agilent | 1260 | Multi-compenent |
| Mass spectrometer | AB Sciex | 4000 |
References
- Sacchettini, J. C., Rubin, E. J., Freundlich, J. S. Drugs versus bugs: in pursuit of the persistent predator Mycobacterium tuberculosis. Nat Rev Microbiol. 6 (1), 41-52 (2008).
- Zhang, Y. Persistent and dormant tubercle bacilli and latent tuberculosis. Front Biosci. 1 (9), 1136-1156 (2004).
- Aber, V. R., Nunn, A. J. Short term chemotherapy of tuberculosis. Factors affecting relapse following short term chemotherapy. Bull Int Union Tuberc. 53 (4), 276-280 (1978).
- Chang, K. C., Leung, C. C., Yew, W. W., Ho, S. C., Tam, C. M. A nested case-control study on treatment-related risk factors for early relapse of tuberculosis. Am J Respir Crit Care Med. 170 (10), 1124-1130 (2004).
- Dartois, V. The path of anti-tuberculosis drugs: from blood to lesions to mycobacterial cells. Nature Rev Microbiol. 12 (3), 159-167 (2014).
- Prideaux, B., et al. The association between sterilizing activity and drug distribution into tuberculosis lesions. Nat Med. 21 (10), 1223-1227 (2015).
- Sarathy, J. P., et al. Prediction of Drug Penetration in Tuberculosis Lesions. ACS Infect Dis. 2 (8), 552-563 (2016).
- Waters, N. J., Jones, R., Williams, G., Sohal, B. Validation of a rapid equilibrium dialysis approach for the measurement of plasma protein binding. J Pharm Sci. 97 (10), 4586-4595 (2008).
- Singh, J. K., Solanki, A., Maniyar, R. C., Banerjee, D., Shirsath, V. S. Rapid Equilibrium Dialysis (RED): an In-vitro High-Throughput Screening Technique for Plasma Protein Binding using Human and Rat Plasma. J Bioequiv Availab. 14, 1-4 (2012).
- Liu, X., et al. Unbound drug concentration in brain homogenate and cerebral spinal fluid at steady state as a surrogate for unbound concentration in brain interstitial fluid. Drug Metab Dispos. 37 (4), 787-793 (2009).
- Able, S. L., et al. Receptor localization, native tissue binding and ex vivo occupancy for centrally penetrant P2X7 antagonists in the rat. Br J Pharmacol. 162 (2), 405-414 (2011).
- Subbian, S., et al. Chronic pulmonary cavitary tuberculosis in rabbits: a failed host immune response. Open Biol. 1 (4), 1-14 (2011).
- Via, L. E., et al. Tuberculous Granulomas are Hypoxic in Guinea pigs, Rabbits, and Non-Human Primates. Infect Immun. 76 (6), 2333-2340 (2008).
- Kalvass, J. C., Maurer, T. S. Influence of nonspecific brain and plasma binding on CNS exposure: implications for rational drug discovery. Biopharm Drug Dispos. 23 (8), 327-338 (2002).
- Di, L., Umland, J. P., Trapa, P. E., Maurer, T. S. Impact of recovery on fraction unbound using equilibrium dialysis. J Pharm Sci. 101 (3), 1327-1335 (2012).
- Lenaerts, A. J., et al. Location of persisting mycobacteria in a Guinea pig model of tuberculosis revealed by r207910. Antimicrob Agents Chemother. 51 (9), 3338-3345 (2007).
- Prideaux, B., et al. High-sensitivity MALDI-MRM-MS imaging of moxifloxacin distribution in tuberculosis-infected rabbit lungs and granulomatous lesions. Anal Chem. 83 (6), 2112-2118 (2011).

