A Novel In Vitro Live-imaging Assay of Astrocyte-mediated Phagocytosis Using pH Indicator-conjugated Synaptosomes
Summary
This protocol presents an in vitro live-imaging phagocytosis assay to measure the phagocytic capacity of astrocytes. Purified rat astrocytes and microglia are used along with pH indicator-conjugated synaptosomes. This method can detect real-time engulfment and degradation kinetics and provides a suitable screening platform to identify factors modulating astrocyte phagocytosis.
Abstract
Astrocytes are the major cell type in the brain and directly contact synapses and blood vessels. Although microglial cells have been considered the major immune cells and only phagocytes in the brain, recent studies have shown that astrocytes also participate in various phagocytic processes, such as developmental synapse elimination and clearance of amyloid beta plaques in Alzheimer's disease (AD). Despite these findings, the efficiency of astrocyte engulfment and degradation of their targets is unclear compared with that of microglia. This lack of information is mostly due to the lack of an assay system in which the kinetics of astrocyte- and microglia-mediated phagocytosis are easily comparable. To achieve this goal, we have developed a long-term live-imaging in vitro phagocytosis assay to evaluate the phagocytic capacity of purified astrocytes and microglia. In this assay, real-time detection of engulfment and degradation is possible using pH indicator-conjugated synaptosomes, which emit bright red fluorescence in acidic organelles, such as lysosomes. Our novel assay provides simple and effective detection of phagocytosis through live-imaging. In addition, this in vitro phagocytosis assay can be used as a screening platform to identify chemicals and compounds that can enhance or inhibit the phagocytic capacity of astrocytes. As synaptic pruning malfunction and pathogenic protein accumulation have been shown to cause mental disorders or neurodegenerative diseases, chemicals and compounds that modulate the phagocytic capacity of glial cells should be helpful in treating various neurological disorders.
Introduction
Glial cells, which refer to non-excitable cells in the brain, are the major cell type in the central nervous system (CNS). Previously, glial cells were regarded as mere supporting cells that mainly play passive roles in maintaining neuronal survival and basal synaptic properties. However, emerging evidence has revealed that glial cells play more active roles in various aspects of neurobiology, such as maintaining brain homeostasis, mediating synapse formation1,2,3 and synapse elimination4,5, and modulating synaptic plasticity6,7. Glial cells in the CNS include astrocytes, microglia, and oligodendrocytes. Among these cells, astrocytes and microglia have been shown to play phagocytic roles by engulfing synapses4,5, apoptotic cells8, neural debris9, and pathogenic proteins, such as amyloid beta plaques10,11. In the developing brain, astrocytes eliminate synapses in the dorsal lateral geniculate nucleus (dLGN) through MERTK- and MEGF10-dependent phagocytosis4. Similarly, microglia also eliminate C1q-coated synapses during developmental stages through the classical complement cascade5. Interestingly, it has been suggested that defects in synapse pruning can be one of the initiators of several neurological disorders. For example, it has been shown that mutations in complement component 4 (C4), which increases complement-mediated synapse pruning by microglia, are strongly associated with the prevalence of schizophrenia in humans12. A recent paper has also shown that the classical complement pathway is hyperactivated in the initiation stages of AD and induces early synapse loss in this disease13.
Compared with microglia-mediated phagocytosis, whether astrocyte-mediated phagocytosis contributes to the initiation and progression of various neurological disorders is less clear. However, a recent paper suggests that factors that alter the rate of normal synapse pruning by astrocytes may disrupt brain homeostasis and contribute to AD susceptibility and pathology14. The rate of synapse pruning by astrocytes is powerfully controlled by ApoE isomers, with a protective allele for AD (ApoE2) strongly enhancing the rate and a risk allele for AD (ApoE4) significantly lowering the rate. Moreover, transgenic mice expressing ApoE4 accumulated much more synaptic C1q than control or ApoE2 mice14. These data suggest that impaired astrocyte-mediated phagocytosis in the early AD brain may induce the accumulation of senescent C1q-coated synapses/synaptic debris that activates complement-mediated microglial phagocytosis, driving synaptic degeneration. The impaired phagocytic capacity of astrocytes in ApoE4 carriers may also contribute to the uncontrolled accumulation of amyloid beta plaques in AD-affected brains.
In addition, it has been shown that glial cells in the aged Drosophila brain lose their phagocytic capacity due to decreased translation of Draper, a homolog of Megf10 that astrocytes use for phagocytosing synapses. Restoring Draper levels rescued the phagocytic capacity of glial cells, which efficiently cleared damaged axonal debris in the aged brain to a similar extent as that in the young brain, indicating that aging-induced alterations in the phagocytic capacity of astrocytes may contribute to disruption of brain homeostasis15.
Based on these new findings, modulating the phagocytic capacity of astrocytes may be an attractive therapeutic strategy to prevent and treat various neurological disorders. In this regard, there have been several attempts to enhance the phagocytic capacity of astrocytes, for example, by inducing acidification of lysosomes with acidic nanoparticles16 and overexpressing transcription factor EB (TFEB), which can enhance lysosome biogenesis17. Despite these attempts, it is still unclear how astrocytes and microglial cells differ in their phagocytic kinetics and whether we should increase or decrease their phagocytic capacities in various diseases.
In this paper, we present a novel in vitro assay for detecting the phagocytic capacity of astrocytes in real-time. The data show different kinetics of engulfment and degradation in astrocytes and microglia. Astrocyte-conditioned medium (ACM), which contains secreted factors from astrocytes, is essential for effective phagocytosis of both astrocytes and microglia. Furthermore, Megf10, a phagocytic receptor in astrocytes and a homolog of Ced-1 and Draper, plays critical roles in astrocyte-mediated phagocytosis8,18.
Protocol
All methods described here have been approved by The Korea Advanced Institute of Science and Technology Institutional Animal Care and Use Committee (IACUC), KA2016-08.
1. Synaptosome Purification
NOTE: These procedures are adapted from a previously published paper19 with several modifications to improve the yield of purified synaptosomes (Figure 1).
- Prepare discontinuous density gradient media.
- Place the following on ice: 3%, 10%, and 23% density gradient solutions in gradient buffer (bring to 250 mL of water with 109.54 g of sucrose, 606 mg of Tris, and 5 mL of 0.2 M EDTA, pH 7.4); homogenizing buffer (add 25 mL of 4x gradient buffer and 500 µL of 50 mM DTT into 74.5 mL of water); homogenizer mortar and pestle.
NOTE: Density gradient solution and preparation are presented in the Table 1. To prepare 3%, 10%, and 23% density gradient solutions, use raw density gradient solution. - Prepare six ultra-clear centrifuge tubes per three adult mice.
- Add 3 mL of 23% density gradient solution on the bottom of each centrifuge tube with a 1-mL pipette. Then, slowly make a layer with 3 mL of 10% density gradient solution on top of the 23% density gradient solution with a pasture pipette and 1-mL pipette. Finally, add 3 mL of 3% density gradient solution on top. In this step, use caution to avoid introducing bubbles.
- Cover the centrifuge tubes with plastic wrap and keep the tubes on ice.
- Place the following on ice: 3%, 10%, and 23% density gradient solutions in gradient buffer (bring to 250 mL of water with 109.54 g of sucrose, 606 mg of Tris, and 5 mL of 0.2 M EDTA, pH 7.4); homogenizing buffer (add 25 mL of 4x gradient buffer and 500 µL of 50 mM DTT into 74.5 mL of water); homogenizer mortar and pestle.
- Precool a high-speed centrifuge and ultracentrifuge to 4 °C.
- Sterilize surgical equipment (operating scissors, Castroviejo spring scissors, and curved forceps) with 70% ethanol. Prepare a small glass beaker with 25 mL of homogenizing buffer on ice and weigh the beaker.
- Extract the brains from three adult (approximately 12 weeks old) male mice.
- Dislocate the cervical vertebra and cut the neck with operating scissors. Peel off the skin of the head and skull along the midline using operating scissors and curved forceps. Excise the olfactory bulbs and cerebellum with Castroviejo spring scissors.
- Put the remaining brains into the beaker with curved forceps and weigh the brains. Rinse the brains several times with ice-cold homogenizing buffer to remove blood on the surface of brains.
NOTE: Previously, 9 mL of homogenizing buffer to 1 g of brain tissues was recommended. - Add 4 mL of homogenizing buffer and 1 g of brain tissues to the homogenizer mortar. While homogenizing the brains, add homogenizing buffer up to 8 mL.
- Divide the homogenate into two 50-mL high-speed centrifuge tubes. Wash the mortar with 1 mL of fresh homogenizing buffer and add it to the tubes.
Caution: Proceed through steps 1.3-1.7 as quickly as possible. Less than 20 min is recommended. - Centrifuge the homogenates in high-speed centrifuge tubes at 1,000 x g for 10 min at 4 °C with slower brakes.
NOTE: Set the deceleration rate (brake) at two or three when the maximum rate of deceleration is nine. - Carefully add the supernatant to a new 50-mL tube and add up to 12 mL of homogenizing buffer.
- Carefully and slowly pipette 2 mL of diluted supernatant over the 3% density gradient solution with a 1-mL pipette and place the tubes on ice.
- Centrifuge at 31,000 x g for 5 min at 4 °C with no brakes.
NOTE: Set the deceleration rate at one (zero) or coast. - Place the tubes on ice and collect the synaptosome fraction between the 23% density gradient solution and 10% density gradient solution with a 2-mL pipette into a 50-mL tube. Add ice-cold sucrose/EDTA buffer up to 80 mL and divide it into four 50-mL high-speed centrifuge tubes.
NOTE: See Figure 3 for fractionated synaptosomes with labeled gradients. - Centrifuge at 20,000 x g for 30 min at 4 ˚C with fewer breaks.
NOTE: Set the deceleration rate at two or three when the maximum rate of deceleration is nine. - Carefully remove the supernatant with a 1-mL pipette, resuspend the synaptosome pellet with 1 mL of isotonic physiological buffer (140 mM NaCl, 3 mM KCl, 1.2 mM MgCl2, 1.2 mM NaH2PO4, 10 mM HEPES, and 10 mM glucose, pH 7.4)20. Add 1 mL of 10% DMSO in isotonic physiological buffer (final concentration of DMSO: 5%) and collect the synaptosomes into one 50-mL high-speed centrifuge tube.
- Aliquot 1 mL into 1.5-mL tubes. Measure the protein concentration of the synaptosomes using the Bradford assay.
- Quickly freeze the tubes and store the tubes in a -80 °C deep freezer until pH indicator conjugation.
2. pH Indicator Conjugation
- Centrifuge 1.5-mL tubes with synaptosomes at 21,092 x g for 3-4 min at 4 °C.
- Remove the supernatant, add 200 µL of 0.1 M Na2CO3 and mix well by pipetting.
NOTE: Na2CO3 was added to raise the pH as succinimidyl ester most efficiently reacts with primary amines at a slightly alkaline pH. - Add 2 µL of pH indicator to the tube and gently vortex.
Caution: Use 1 µL of pH indicator per 0.3 mg of synaptosome solution. - Minimize light exposure by covering the tubes with aluminum foil and incubate them for 1-2 h at room temperature (RT) in a twist shaker with gentle agitation at 30-40 rpm.
- Stop agitation and add 1 mL of DPBS.
- Centrifuge the tubes at 21,092 x g for 1-2 min.
- Remove the supernatant and add 1 mL of DPBS to resuspend the pellet by gentle pipetting.
- Repeat steps 2.6-2.7 at least seven times to remove unbound pH indicator.
- Remove the supernatant and resuspend the pellet with 200 µL of DPBS with 5% DMSO.
NOTE: At this point, all synaptosomes are non-functional. We have confirmed that phosphatidylserine (PS) has been exposed to the outer membrane of synaptosomes, which functions as an "eat-me" signal (Figure 4).
3. Astrocytes and Microglia Purification
NOTE: This protocol for astrocyte purification is adapted from a previously published paper21. In this protocol, the purification of rat astrocytes and microglia is described. Specific procedures for purifying mouse astrocytes and microglia are also described in the notes.
- Day before purification
- Prepare pre-coated cell culture dishes and solutions.
- Prepare five 145 mm x 20 mm Petri dishes with 25 mL of 50 mM Tris-HCl (pH 9.5). Place primary or secondary antibody treatment in the Petri dishes separately as described below. Incubate the dishes in the freezer overnight.
BSL-1 dish: 20 µL of 5 mg/mL BSL-1 (Griffonia simplicifolia lectin)
Secondary-only dish: 60 µL of 2.4 mg/mL goat anti-mouse IgG+IgM (H+L)
CD45 dish: 60 µL of 2.4 mg/mL goat anti-mouse IgG+IgM (H+L)
O4 dish: 60 µL of 2.4 mg/mL goat anti-mouse IgM (µ-chain specific)
Itgb5 (Integrin β5) dish for rat astrocytes: 60 µL of 2.4 mg/mL goat anti-mouse IgG+IgM (H+L)
NOTE: To attach secondary antibody to the hydrophobic surface of the Petri dish, slow attachment at low temperatures is recommended. However, it is possible to coat the Petri dish with secondary antibody for 2 h at 37 °C. Antibodies for astrocyte panning are used differently depending on the animal species; for example, HepaCAM dish for mouse astrocytes: 60 µL of 2.4 mg/mL goat anti-mouse IgG+IgM (H+L)
- Day of Purification
- Prepare for immunopanning.
- Prepare 50-mL tubes and add stocks to the tubes. The detailed stock solution preparation is presented in the Table 1.
- Prepare Tube 1 (Enzyme): 22 mL of enzyme stock (Enzyme stock: 1x Earle's Balanced Salt Solution (EBSS)+0.46% D(+)-glucose+26 mM NaHCO3 and0.5 mM EDTA).
- Prepare Tube 2 (Low Ovo): 42 mL of inhibitor stock (Inhibitor stock: 1x EBSS+0.46% D(+)-glucose+26 Mm NaHCO3).
- Prepare Tube 3 (High Ovo): 10 mL of inhibitor stock.
- Attach 0.22-µm syringe filters to the tips of 2-mL pipettes connected to a CO2 (95% O2 and 5% CO2) tank. Bubble CO2 into Tubes 1-3. Stop bubbling when the solutions turn orange.
- Complete the solutions.
Caution: Complete and incubate the solution in Tube 1 at 34 °C for 15 min before using for enzyme activation.- Prepare Tube 1 (Enzyme): 22 mL of enzyme stock + 180 U of papain + 0.004 g L-cysteine.
- Prepare Tube 2 (Low Ovo): 42 mL of inhibitor stock + 3 mL of Low Ovo + 200 µL of DNase.
- Prepare Tube 3 (High Ovo): 10 mL of inhibitor stock + 2 mL of High Ovo + 20 µL of DNase.
- Prepare Tube 4 (0.2% BSA): 19 mL of 1X DPBS + 1 mL of 4% BSA + 8 µL of DNase.
- Prepare Tube 5 (0.02% BSA): 45 mL of 1X DPBS + 5 mL of Tube 4 + 50 µL of DNase.
- Preheat a water bath and heat block at 34 °C.
- Extract the rat cortex.
- Sterilize surgical equipment with 70% ethanol and prepare 100-mm Petri dishes with DPBS.
- Prepare an acrylic box. Put two to three layers of tissues in the bottom of the box and add 1 mL of isoflurane into the box.
- Anesthetize pups for 20-40 s in the box and confirm anesthetization by foot pinch with forceps. Expose the heart22 and perfuse pups using a 10-mL syringe with DPBS.
NOTE: Perfusion is used to avoid blood cell contamination during the microglia panning step. - Cut the pup's upper cervical line of the neck with operating scissors and incise the head skin along the midline. Make an incision on the skull along the midline and open the skull with curved forceps.
- Excise the cerebellum with Castroviejo spring scissors. Take out the remaining brain and place it in a 100-mm Petri dish filled with DPBS.
- Remove other areas such as the midbrain and hippocampus from the cortex with Castroviejo spring scissors under the microscope. Remove the meninges with fine forceps.
- Transfer the cortex into a new 60-mm dish and remove the remaining DPBS in the dish. Chop the tissue with a No. 10 blade.
- Dissociate the cortex into a single cell suspension
- Pour the filtered Tube 1 solution into a 60-mm dish and add 50 µL of DNase1. Swirl the solution in the dish.
NOTE: DNase is used to inhibit aggregation of DNA from ruptured cells. - Place the dish into a 34 °C heat block and cover it with a lid that has a hole in the middle. Use a soldering iron to make the hole in the lid of the 60-mm Petri dish.
- Connect 22-µm syringe filters to a CO2 tank tube and place the filter tip into the hole.
- Incubate the dish at 34 °C for 45 min. Swirl the solution in the dish every 10 to 15 min.
- Pour the filtered Tube 1 solution into a 60-mm dish and add 50 µL of DNase1. Swirl the solution in the dish.
- Finish preparation of the panning dishes.
- Remove the panning dishes that have been coated with secondary antibodies from the freezer a day ahead of time. Leave the BSL-1 dish and secondary antibody-only dish at RT.
- Wash the other dishes three times with 20 mL of DPBS.
- Pour an adequate amount of 0.02% BSA into the dishes. Place specific primary antibodies into the corresponding dishes:
CD45 dish: 12 mL of 0.02% BSA + 20 µL of 0.5 mg/mL Rat anti-mouse CD45
Itgb5 dish: 12 mL of 0.02% BSA + 20 µL of 0.5 mg/mL Itgb5
O4 dish: 8 mL of 0.02% BSA and 4 mL of O4 antibody
NOTE: For mouse immunopanning, use mouse anti-rat CD45 (0.5 mg/mL) for microglia and HepaCAM (0.5 mg/mL) for astrocytes. - Prepare 30 mL of 30% FBS with neurobasal media and 10 mL of 1X EBSS. Warm up both solutions in a 34 °C water bath.
- Inhibit the papain reaction.
- Transfer the papain-treated tissues to a new 50-mL tube. Wait for the tissues to settle. Aspirate liquid by suction.
- Add 4 mL of Low Ovo solution to the tube and swirl the solution to wash the cells.
- Repeat steps 3.6.1-3.6.2 three times to stop the enzyme reaction.
- Triturate the cortex tissues.
- Add 6 mL of Low Ovo to the tub. Aspirate and release the tissues quickly with a 5-mL pipette.
Caution: Do not introduce bubbles. - Prepare a new 50-mL tube and add 5 mL of Low Ovo. Collect single cells in a new tube.
- Repeat steps 3.6.1-3.6.2 two times and change the 5-mL pipette to a 1-mL pipette.
- Repeat trituration until more than 95% of the tissues are not visible.
- Make a layer of High Ovo solution from tube 3 with a 10-mL pipette below low Ovo-containing dissociated cells.
- Centrifuge at 190 x g for 6 min with few brakes.
NOTE: Set the deceleration rate at one or two when the maximum rate of deceleration is nine.
- Add 6 mL of Low Ovo to the tub. Aspirate and release the tissues quickly with a 5-mL pipette.
- Filtrate single cells.
- Carefully aspirate the supernatant by suction, then resuspend the pellet with 3 mL of 0.02% BSA solution with a 1-mL pipette.
- Add 0.02% BSA solution up to 9 mL.
- Prepare a new 50-mL tube and 20-µm cell strainer on top of the tube. Wet the cell strainer with 1 mL of 0.02% BSA solution.
- Transfer the single cell suspension into the cell strainer. Filter 1 mL at a time.
- Wash the cell strainer with 3 mL of 0.02% BSA solution. Do not make more than 15 mL of the filtered single cell suspension.
- Incubate the tube in a 34 °C water bath for 45-60 min for cell recovery. The cell recovery step allows for relocalization of cell surface proteins to the membrane.
- Perform panning of the cells.
- Wash the CD45 panning dish three times with 20 mL of DPBS. Wash every dish three times with 20 mL of DPBS immediately before use.
- Pour the single cell suspension into the CD45 panning dish. Leave the dish for 28 min and shake the dish for 14 min. To remove unbound cells from the bottom, carefully shake the dish.
- Transfer the cell suspension to the secondary-only dish. Wait for 20 min and carefully shake the dish for 10 min. To obtain microglia from the CD45 dish, go to step 3.9.2.
- Transfer the cell suspension into the BSL-1 dish. Leave the dish for 10 to 12 min.
- Transfer into the O4 dish. Wait for 20 min and shake for 10 min.
- Transfer into the Itgb5 dish. Leave the dish for 40 min and gently shake every 10 min.
NOTE: For mouse panning, transfer the cell suspension into the HepaCAM dish.
- Detach cells from the dish.
- Mix 400 µL of trypsin (30,000 U/mL in EBSS) with preincubated 8 mL of 1X EBSS.
- Pour the detached cell suspension in the dish into a waste bucket.
- Wash the dish eight times with DPBS.
- Pour 8 mL of EBSS with trypsin into the dish.
- Incubate the dish for 5 to 10 min in the 37 °C incubator. Incubate for 10 min for the CD45 dish and 5 min for the Itgb5 and HepaCAM dishes.
- Tap the dish and observe the dish under a microscope.
NOTE: If most of the astrocytes have not detached, put the dish back into the incubator for another 5 min. - Pour 10 mL of 30% FBS into the dish to neutralize trypsin.
- Pipette up and down in the entire dish to detach astrocytes or microglia.
- Transfer the solution into a 50-mL tube.
- Add 10 mL of 30% FBS and 200 µL of DNase into the dish. Detach the cells again. DNase is used to inhibit aggregation of DNA from ruptured cells.
- Transfer the solution into the tube. If cells remain in the dish, add another 10 mL of 30% FBS and detach the cells again.
- Plate the cells.
- Centrifuge the tube at 213 x g for 10 min.
- Aspirate the supernatant and resuspend the cell pellet with media.
NOTE: Use immunopanned astrocyte basic media (IP-ABM) for astrocytes and retinal microglia basic media (MBM) with immunopanned astrocyte-conditioned media (IP-ACM) for microglia. The IP-ABM and MBM compositions are presented in the Table of Materials. - Count the cells with a hemocytometer.
- Plate the astrocytes and microglia separately in PDL-coated plates.
NOTE: Prepare the PDL-coated plate before use. To prepare PDL-coated plates, add 50 µL of 1 mg/mL poly-D-lysine into 5 mL of distilled water and pour an adequate amount of solution into the well plate/dish, and wait for 30 min. Wash the plate/dish three times with distilled water. - Incubate the plate. Change half of the media every 7 days for astrocytes and 3 days for microglia.
4. Collect IP-ACM
- Prepare a 100-mm dish when astrocytes are overly confluent.
- Discard the media and wash the dish three times with 10 mL of DPBS.
- Pour 15 mL of low protein media into the dish.
NOTE: The low protein media composition is presented in the Table of Materials. - Incubate the dish for 7 to 10 days in the 37 °C incubator.
- Collect and concentrate the media with 10k (or 30k) centrifugal filter tubes.
- Centrifuge the tubes at 851 x g at 4 °C.
NOTE: Centrifuge the tubes until the remaining media is less than 1 mL. - Collect concentrated media (IP-ACM) into a new 1.5-mL tube.
- Measure the concentration of IP-ACM using quantitative analysis
5. Phagocytosis Live-imaging Assay (Figure 2)
- Prepare a 24-well plate (or any plate/dish that is prepared for live-imaging) in which astrocytes are confluent (generally, 7 to 10 days of incubation after the purification is ideal for stabilizing the cells).
- Remove the media in each well and wash the wells with 1 mL of DPBS, three times. To minimize air exposure, wash the wells quickly.
- Add 300 µL of IP-ABM with 5 µL of pH indicator-conjugated synaptosomes and additional factors such as IP-ACM that can modulate glial cell phagocytosis.
- Incubate the plate in a CO2 incubator for 40 min. This step allows the pH indicator-conjugated synaptosomes to settle to the bottom of the plates.
- Remove the media with unbound pH indicator-conjugated synaptosomes and wash each well with 1 mL of DPBS, three times.
Caution: Do not wash harshly. To minimize air exposure, wash the wells quickly. - Add 500 µL of IP-ABM with additional factors to test into the wells.
- Take the plate to a live-imaging instrument and select positions. Adjust focus, exposure time, brightness, and LED power.
NOTE: The settings for exposure time, brightness, and LED power are variable depending on fluorescence intensity and the purpose of the experiment. In the live-imaging assay with pH indicator-conjugated synaptosomes, we usually set those values as follows: exposure: ~ 150-200 ms, brightness: 15, and LED power: 4-6. - Set the image format, time interval, and the total number of cycles for live-imaging. Set the time interval to 1 or 2 h depending on how many positions are selected. 2 hour intervals are recommended when more than 150 positions are selected for live-imaging.
- Start the live-imaging experiments.
- Analyze the data.
- Open the Fiji program.
- Import an image sequence. Convert images to 8-bit grayscale.
- Perform background subtraction with a rolling bar radius of 50 pixels.
- Start time series analyzer V3 plugins.
NOTE: The address for time series analyzer V3 plugins is presented in the Table of Materials. - Drag the region of interest (ROI) in the image and click add in the ROI manager.
- Click "Get total intensity" in the Time series V3_0.
- Save the results and integrate the data.
Representative Results
In this in vitro phagocytosis assay with long-term live-imaging, we used synaptosomes from adult mouse brain homogenates, which were separated in the gradient solution between 23% gradient solution and 10% gradient solution by ultracentrifugation (Figure 3). After the preparation, synaptosomes exposed PS in their outer membrane (Figure 4), suggesting that they lost their function and could be recognized by PS receptors in astrocytes and microglia. As shown in Figure 5, pH indicator-conjugated synaptosomes emitted bright red fluorescence when they were engulfed by astrocytes. Real-time comparison of the engulfment and degradation capacities of glial cells is possible by taking multiple images of a ROI every 1 or 2 h (Supplementary Movie 1, Supplementary Movie 2). With this method, we demonstrated different kinetics of astrocyte- and microglia-mediated phagocytosis (Figure 6). To quantify phagocytosis of glial cells, the area (µm2) of red fluorescence signal was measured, which is referred to phagocytic index in Figure 6B and Figure 7. Although astrocytes appeared to be efficient in phagocytosing large amounts of pH indicator-conjugated synaptosomes, microglia were faster at engulfing and degrading synaptosomes (Figure 6B). Microglia showed maximum pH indicator intensity at 26 h after pH indicator-conjugated synaptosome treatment, whereas astrocytes showed their maximum at 45 h (p-value < 0.05, two-way ANOVA between astrocyte with 1X ACM and microglia with 1X ACM). Likewise, microglial cells showed a 20.7% reduction in total pH indicator intensity 40 h after the peak point, whereas astrocytes showed a 17% reduction in total pH indicator intensity during the same time period. Interestingly, our data showed that astrocyte-secreted factors, which were contained in ACM, are essential for increasing both astrocyte- and microglia-mediated phagocytosis (Figure 6B). Astrocytes have been shown to release bridging molecules, such as MFGE8, GAS6, and ProteinS, which can bridge and induce interactions between phagocytic receptors and "eat-me" signals such as PS23. As mentioned above, astrocytes eliminate synapses via the MERTK and MEGF10 pathways4. MEGF10 is only expressed by astrocytes in the brain and participates in synapse engulfment through recognizing "eat-me" signals with unknown identities. In agreement with previous findings, the assay showed that compared with wild-type (WT) mouse astrocytes, Megf10 knock-out (KO) mouse astrocytes possessed significantly impaired phagocytic capacity (Figure 7). Compared with WT astrocytes, Megf10 KO astrocytes showed an approximately 40% reduction in total pH indicator intensity at the peak point (31 h) (Figure 7).
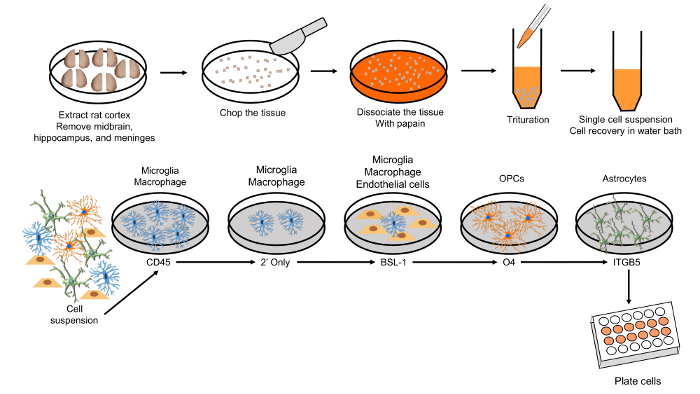
Figure 1. Schematic of astrocyte purification using immunopanning methods. Please click here to view a larger version of this figure.
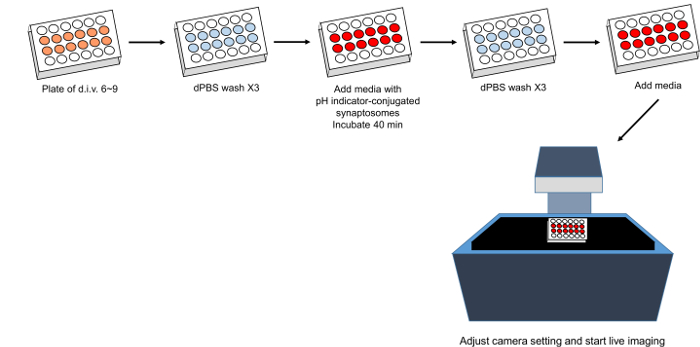
Figure 2. Schematic of phagocytosis live-imaging assay. Please click here to view a larger version of this figure.
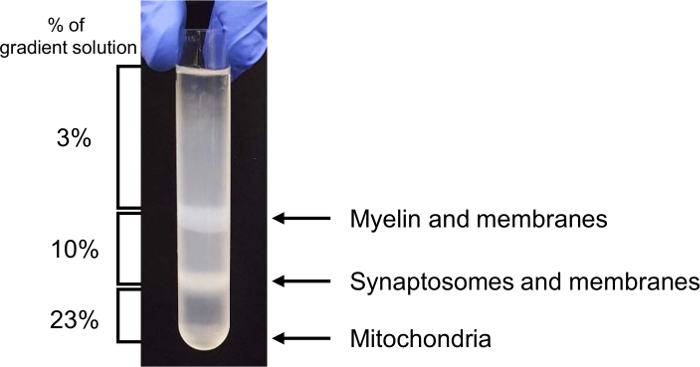
Figure 3. Representative brain homogenate fractionation in the gradient solutions. Please click here to view a larger version of this figure.
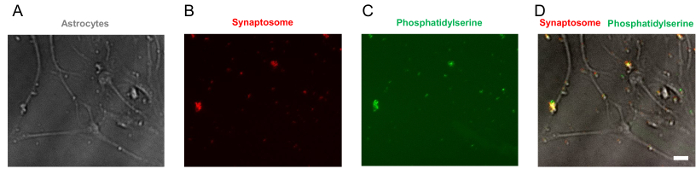
Figure 4. Representative images of PS-exposed synaptosomes. (A) A bright field image of astrocytes with synaptosomes. Synaptosomes are attached to astrocytes. (B) A fluorescent image of tdTomato-positive synaptosomes, which are purified from tdTomato-expressing mouse brains. (C) pSIVA binds to PS, which is exposed to outer membrane of synaptosome, and emits green fluorescence. (D) PS detected by pSIVA (green) is co-localized with tdTomato-positive synaptosomes (red). Scale bar: 20 µm Please click here to view a larger version of this figure.
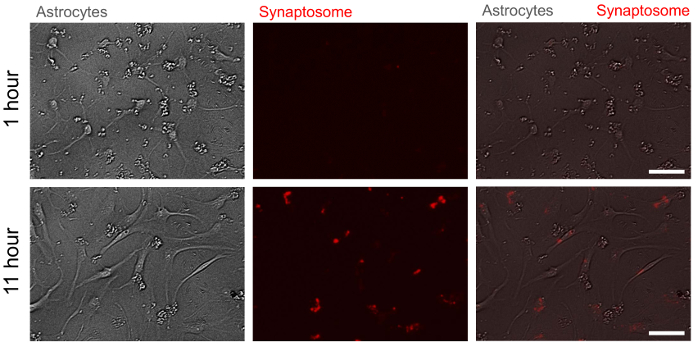
Figure 5. Representative bright field and fluorescent images of astrocytes with pH indicator-conjugated synaptosomes at two time points. At 11 h after treatment (bottom panel), pH indicator-conjugated synaptosomes are engulfed by astrocytes and emit red fluorescence whereas they do not at 1 h after treatment (upper panel). Scale bar: 50 µm. Please click here to view a larger version of this figure.
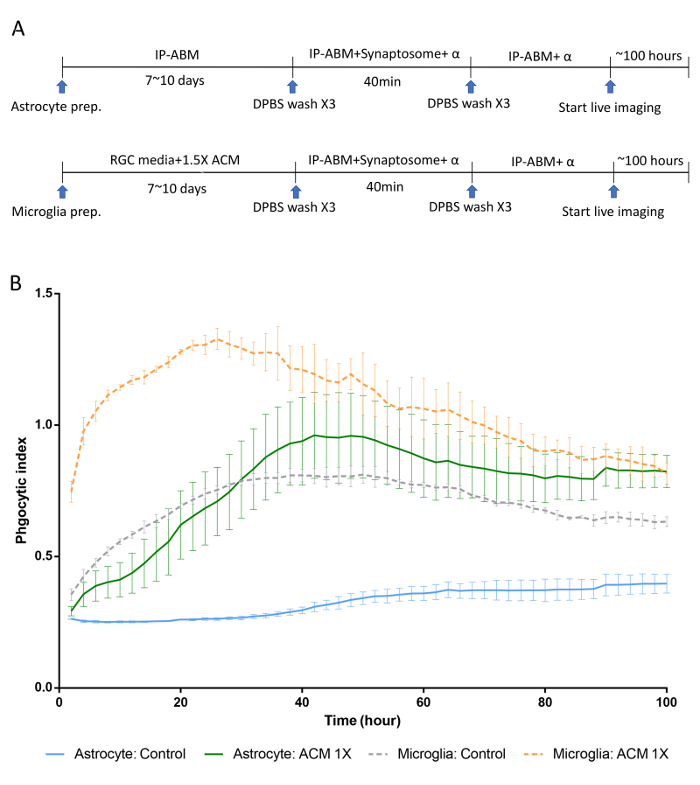
Figure 6. Phagocytic kinetics of rat astrocytes and microglia through long-term live-imaging. (A) A schematic diagram of an in vitro phagocytosis assay using purified astrocytes and microglia along with pH indicator-conjugated synaptosomes. Note that before live images are taken, unbound synaptosomes are washed away after 40 min of incubation. (B) Representative graphs showing the engulfment and degradation kinetics of astrocytes and microglia. ACM, which contains astrocyte-secreted factors, significantly enhances both astrocyte- and microglia-mediated synaptosome uptake. Astrocyte: Control vs. Astrocyte with ACM 1X, ****, Tukey's multiple comparisons test. Microglia: Control vs. Microglia with ACM 1X, ****, Tukey's multiple comparison test. Error bars indicate S.E.M. *p ≤ 0.05, **** p ≤ 0.0001, two-way ANOVA. Please click here to view a larger version of this figure.
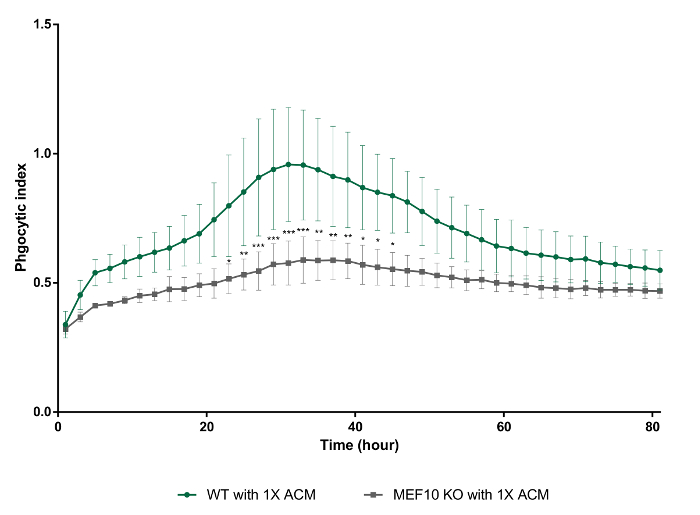
Figure 7. Decreased phagocytic capacity of Megf10 KO mouse astrocytes with 1X ACM compared with that of WT mouse astrocytes with 1X ACM. WT vs. Megf10 KO astrocytes with 1X ACM, ****, two-way ANOVA. Error bars indicate S.E.M. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001 two-way ANOVA. Please click here to view a larger version of this figure.
Table 1: Solution recipes. Please click here to download this file.
Supplemental Movie 1. A representative live-imaging video showing the phagocytosis of pH indicator-conjugated synaptosomes by astrocytes. Please click here to download this file.
Supplemental Movie 2. A representative live-imaging video showing phagocytosis of pH indicator-conjugated synaptosomes by microglial cells. Please click here to download this file.
Discussion
In this article, we present methods for a long-term live-imaging in vitro phagocytosis assay using purified glial cells and pH indicator-conjugated synaptosomes. We show that compared with microglia, astrocytes possess different engulfment and degradation capacity during the phagocytosis of synaptosomes. In addition, our data suggest that astrocyte-secreted factors, which contain bridging molecules such as GAS6, ProteinS, and MEGE8, are essential for efficient PS-dependent phagocytosis of glial cells in the brain. Furthermore, Megf10 KO astrocytes show defective phagocytosis of synaptosomes.
To successfully perform this experiment, the media and DPBS need to be carefully exchanged during the washing steps (Section 5). Primary cultured astrocytes and microglia, especially microglia, are vulnerable to air exposure. Therefore, reducing time intervals between washing steps is critical for maintaining live cells. For setting up long-term live-imaging with live-imaging instruments, manual focus is recommended since auto focus may increase laser/LED exposure time, which could damage the cells.
In this method, the synaptosome purification protocol19 is slightly modified. The original paper separates gradient solutions into 3%, 10%, 15%, and 23%. However, we set up 3%, 10% and 23% gradient solutions to increase the yield of purified synaptosomes. The instructions for immunopanning astrocytes and microglial cells are also modified in this method. The goal of the original panning protocol21 is to purify only astrocytes and use BSL-1, secondary-only, CD45, O4, and Itgb5 (HepaCAM for mouse) as the immunopanning order. Since BSL-1 and secondary-only plates will remove endothelial cells as well as microglia, we changed the order to CD45, secondary-only, BSL-1, O4, and Itgb5 (HepaCAM for mouse) to increase the yield of microglia from the CD45 panning dish. In this protocol, we also perfused SD rat pups (~ P7-P10) with DPBS through the circulatory system to remove various blood cells to minimize contamination from CD45-positive non-microglial populations.
There are several advantages of the in vitro phagocytosis assay. The pH indicator used in synaptosome conjugation only emits red fluorescence in low pH conditions. Therefore, when processing data, we can directly quantify red fluorescence as a signal of phagocytic events without a quenching procedure to remove background signals or complicated imaging analysis to obtain co-localized signals of engulfed material inside of phagocytes. In addition, this method allows real-time tracking of glial phagocytosis by taking images of pH indicator intensity within a given ROI at multiple time points. By generating a graph with pH indicator intensity up to 100 h, the engulfment as well as degradation capacities can be easily monitored between different cells and conditions. Another advantage is the use of synaptosomes. Since astrocytes ensheathe synapses all the time with their fine processes and have been shown to engulf synapses in vivo4, using synaptosomes is very suitable for measuring the in vitro phagocytic capacity of astrocytes. Finally, this in vitro phagocytosis assay has the potential to be developed to study glial phagocytosis of myelin debris or amyloid beta, which is related to various neurological disorders.
With increased life expectancy, a dramatic increase in the number of patients with neurodegenerative diseases is inevitable. Synapse loss through glial cells is one of the leading factors in several neurodegenerative diseases13,14. In addition, abnormal synapse pruning and an imbalance of brain homeostasis can be associated with glial phagocytosis defects. Therefore, identifying factors and compounds that can control the engulfment and degradation capacities of astrocytes could be critical for finding successful treatments for various neurological disorders. Since this in vitro phagocytosis assay can be easily scaled up with multiwell plates, this method can be used as a suitable platform for various screenings to identify such factors.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank Yeon-Joo Jung for her experimental support during synaptosome purification and Jungjoo Park for images of synaptosomes with PS exposure. In addition, we thank all members in Chung's laboratory for helpful discussion. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (NRF-2016M3C7A1905391 and NRF-2016R1C1B3006969) (W.-S. C).
Materials
| Synaptosome purification | |||
| Percoll | GE healthcare life sciences | 17-0891-01 | |
| Quick Start Bradford Protein Assay Kit 2 | BIO-RAD | 5000202 | |
| pH indicator conjugation | |||
| Dimethyl sulfoxide(DMSO) | LPS solution | DMSO100 | |
| pHrodo red, succinimidyl ester | Molecula probes | P36600 | |
| Immunopanning | |||
| 10X Earle’s balanced salt solution (EBSS) | Sigma | E7510 | |
| Bovine serum albumin | Bovogen | BSA025 | |
| Deoxyrebonuclease 1 (DNase) | Worthington | Is002007 | |
| (DMEM) | Gibco | 11960-044 | |
| (dPBS) | Welgene | LB001-02 | |
| Fetal bovine serum (FBS) | Gibco | 16000-044 | |
| Griffonia Simplicifolia Lectin(BSL-1) | Vector Labs | L-1100 | |
| Goat anti-mouse IgG+IgM(H+L) | Jackson ImmunoResearch | 115-005-044 | |
| Goat anti-mouse IgM (μ-chain) | Jackson ImmunoResearch | 115-005-020 | |
| Heparin-binding epidermal growth factor | Sigma | E4643 | |
| Human HepaCAM antibody | R&D systems | MAB4108 | |
| Integrin beta 5 monoclonal antibody (KN52) | eBioscience | 14-0497-82 | |
| L-cysteine | Sigma | C7880 | |
| L-glutamate | Gibco | 25030-081 | |
| N-acetly-L-cyteine (NAC) | Sigma | A8199 | |
| Neurobasal media | Gibco | 21103-049 | |
| O4 hybridoma supernatant(mouse IgM) | Bansal et al.23 | ||
| Papain | Worthington | Is003126 | |
| Penicillin/streptomycin | Gibco | 15140-122 | |
| Pluristrainer 20 μm | PluriSelect | 43-50020-03 | |
| Poly-D-lysine | Sigma | P6407 | |
| Progesterone | Sigma | P8783 | |
| Putrescine dihydrochloride | Sigma | P5780 | |
| Purified rat anti-mouse CD45 | BD Pharmingen | 550539 | |
| Purified mouse anti-rat CD45 | BD Pharmingen | 554875 | |
| Sodium pyruvate | Gibco | 11360-070 | |
| Sodium selenite | Sigma | S5261 | |
| Transferrin | Sigma | T1147 | |
| Trypsin | Sigma | T9935 | |
| Trypsin inhibitor | Worthington | LS003086 | |
| Ultra-clear tube (Tube, Thinwall, Ultra-Clear) | Beckman Coulter | 344059 | |
| Collect IP-ACM | |||
| Macrosep Advance Centrifugal Devices with Omega Membrane (10k) | PALL | MAP010C37 | |
| Macrosep Advance Centrifugal Devices with Omega Membrane (30k) | PALL | MAP030C37 | |
| Phagocytosis live imaging assay | |||
| Juli stage | NanoEntek | ||
| Time Series Analyzer V3 plugins | https://imagej.nih.gov/ij/plugins/time-series.html |
References
- Singh, S. K., et al. Astrocytes Assemble Thalamocortical Synapses by Bridging NRX1alpha and NL1 via Hevin. Cell. 164 (1-2), 183-196 (2016).
- Allen, N. J., et al. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 486 (7403), 410-414 (2012).
- Xu, J., Xiao, N., Xia, J. Thrombospondin 1 accelerates synaptogenesis in hippocampal neurons through neuroligin 1. Nat Neurosci. 13 (1), 22-24 (2010).
- Chung, W. S., et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 504 (7480), 394-400 (2013).
- Schafer, D. P., et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 74 (4), 691-705 (2012).
- Ma, Z., Stork, T., Bergles, D. E., Freeman, M. R. Neuromodulators signal through astrocytes to alter neural circuit activity and behaviour. Nature. 539 (7629), 428-432 (2016).
- Parkhurst, C. N., et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 155 (7), 1596-1609 (2013).
- Iram, T., et al. Megf10 Is a Receptor for C1Q That Mediates Clearance of Apoptotic Cells by Astrocytes. J Neurosci. 36 (19), 5185-5192 (2016).
- Tasdemir-Yilmaz, O. E., Freeman, M. R. Astrocytes engage unique molecular programs to engulf pruned neuronal debris from distinct subsets of neurons. Genes Dev. 28 (1), 20-33 (2014).
- Jones, R. S., Minogue, A. M., Connor, T. J., Lynch, M. A. Amyloid-beta-induced astrocytic phagocytosis is mediated by CD36, CD47 and RAGE. J Neuroimmune Pharmacol. 8 (1), 301-311 (2013).
- Wang, Y., et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 160 (6), 1061-1071 (2015).
- Sekar, A., et al. Schizophrenia risk from complex variation of complement component 4. Nature. 530 (7589), 177-183 (2016).
- Hong, S., et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 352 (6286), 712-716 (2016).
- Chung, W. S., et al. Novel allele-dependent role for APOE in controlling the rate of synapse pruning by astrocytes. Proc Natl Acad Sci U S A. 113 (36), 10186-10191 (2016).
- Purice, M. D., Speese, S. D., Logan, M. A. Delayed glial clearance of degenerating axons in aged Drosophila is due to reduced PI3K/Draper activity. Nat Commun. 7, 12871 (2016).
- Loov, C., Mitchell, C. H., Simonsson, M., Erlandsson, A. Slow degradation in phagocytic astrocytes can be enhanced by lysosomal acidification. Glia. , (2015).
- Xiao, Q., et al. Enhancing astrocytic lysosome biogenesis facilitates Abeta clearance and attenuates amyloid plaque pathogenesis. J Neurosci. 34 (29), 9607-9620 (2014).
- Lu, T. Y., et al. Axon degeneration induces glial responses through Draper-TRAF4-JNK signalling. Nat Commun. 8, 14355 (2017).
- Dunkley, P. R., Jarvie, P. E., Robinson, P. J. A rapid Percoll gradient procedure for preparation of synaptosomes. Nat Protoc. 3 (11), 1718-1728 (2008).
- Stigliani, S., et al. Glia re-sealed particles freshly prepared from adult rat brain are competent for exocytotic release of glutamate. J Neurochem. 96 (3), 656-668 (2006).
- Foo, L. C., et al. Development of a method for the purification and culture of rodent astrocytes. Neuron. 71 (5), 799-811 (2011).
- Gage, G. J., Kipke, D. R., Shain, W. Whole animal perfusion fixation for rodents. J Vis Exp. (65), (2012).
- Cahoy, J. D., et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 28 (1), 264-278 (2008).

