Visualization of Microbiota in Tick Guts by Whole-mount In Situ Hybridization
Summary
Here, we present a protocol to spatially and temporally assess the presence of viable microbiota in tick guts using a modified whole-mount in situ hybridization approach.
Abstract
Infectious diseases transmitted by arthropod vectors continue to pose a significant threat to human health worldwide. The pathogens causing these diseases, do not exist in isolation when they colonize the vector; rather, they likely engage in interactions with resident microorganisms in the gut lumen. The vector microbiota has been demonstrated to play an important role in pathogen transmission for several vector-borne diseases. Whether resident bacteria in the gut of the Ixodes scapularis tick, the vector of several human pathogens including Borrelia burgdorferi, influence tick transmission of pathogens is not determined. We require methods for characterizing the composition of the bacteria associated with the tick gut to facilitate a better understanding of potential interspecies interactions in the tick gut. Using whole-mount in situ hybridization to visualize RNA transcripts associated with particular bacterial species allows for the collection of qualitative data regarding the abundance and distribution of the microbiota in intact tissue. This technique can be used to examine changes in the gut microbiota milieu over the course of tick feeding and can also be applied to analyze expression of tick genes. Staining of whole tick guts yield information about the gross spatial distribution of target RNA in the tissue without the need for three-dimensional reconstruction and is less affected by environmental contamination, which often confounds the sequencing-based methods frequently used to study complex microbial communities. Overall, this technique is a valuable tool that can be used to better understand vector-pathogen-microbiota interactions and their role in disease transmission.
Introduction
Human and livestock pathogens transmitted by arthropod vectors are found worldwide and account for about 20% of infectious diseases globally1, but effective and safe vaccines against most of these pathogens are not available. Our understanding of the important role of commensal, symbiotic and pathogenic microorganisms, collectively known as the microbiome2, in modulating and shaping the health of almost all metazoans3 is expanding. It is now evident that arthropod vectors of pathogens also harbor gut microbiota and these vector-associated microbiota have been shown to influence diverse vector-borne pathogens4,5. The arthropod microbiome is composed of eubacteria, archaea, viruses, and eukaryotic microbes such as protozoa, nematodes, and fungi6. However, the predominant research focus has been on eubacteria due, in part, to the availability of marker genes and reference databases to identify specific bacterial members.
With a focus on Ixodes scapularis, the tick vector of multiple human pathogens including Borrelia burgdorferi7, the causative agent of Lyme disease, the optimization of a microbial visualization technique was aimed at improving our understanding of tick gut microbiota in the context of vector-pathogen interactions. Several questions remain to be answered in the tick microbiome field. The gut is the site of the first extended encounter between the tick and the incoming pathogen in the context of horizontally transferred pathogens; therefore, understanding the role of vector gut microbiota in modulating vector-pathogen interactions will reveal meaningful insights. Ticks have a unique mode of blood meal digestion, where processing of blood meal components takes place intracellularly8. The gut lumen seemingly serves as a vessel to contain the blood meal as the tick feeds, and nutrient digestion and assimilation ensue throughout the several days of feeding and continue post-repletion. The pathogens acquired by the tick during feeding enter the gut lumen along with the bloodmeal and thus the lumen becomes a primary site of interactions among the tick, pathogen, and resident microbiota. As digestion proceeds through the repletion and Ixodid tick molting, the gut undergoes structural and functional changes9. The composition and the spatial organization of gut bacteria is also likely to vary in concert with the changing gut milieu. It is, therefore, important to understand the architecture of resident bacteria in the tick gut to fully understand the interplay of tick, pathogen, and gut microbiota.
Molecular techniques to describe host-associated microbiota routinely utilize high-throughput parallel sequencing strategies10 to amplify and sequence bacterial 16S ribosomal DNA (rDNA). These sequencing strategies circumvent the need to obtain axenic cultures of specific bacteria and provide an in-depth description of all bacterial members represented in the sample. Nevertheless, such strategies are confounded by the inability to distinguish environmental contaminations from bona fide residents. Further, when assessing samples, such as ticks, that are small in size and hence contain low microbiota-specific DNA yields, the likelihood of amplification of environmental contaminants is increased11 and results in the ambiguous interpretation of microbiome composition. Functional characterization in conjunction with the visualization of specific viable bacteria will, therefore, be critical to define and discern the microbiome of the tick temporally and spatially. Towards this goal, we took advantage of the whole-mount RNA in situ hybridization. This technique is routinely used to assess gene expression patterns in organs and embryos12,13,14 and allows semiquantitative analysis of expression over the entire sample of interest. This differs from traditional in situ hybridization techniques which utilize tissue sections and often require extensive analysis of sectioned material with a computational assembly to predict expression in whole organs15. While whole-mount generally refers to whole organisms12, here whole-mount refers to whole guts or organs. The advantages of using the whole-mount RNA in situ hybridization approach to assess the architecture of tick gut microbiota are multifold. The tick gut is composed of 7 pairs of diverticula, each pair varying in size16. The functional differences, if any, among these diverticula, are not understood in the context of tick biology, tick microbiota or tick-pathogen interactions. Manipulations of the gut that rupture the gut diverticula would displace microbiota present in the gut lumen or those associated loosely with the gut and result in misinterpretation of the spatial localization of microbiota. Fluorescence-labeled RNA in situ hybridization has been utilized earlier to examine tick gut transcripts17 by fixing and opening individual gut diverticula to ensure probe hybridization and to localize B. burgdorferi transcripts by sectioning paraffin-embedded whole ticks18. Both these approaches require manipulations of the tick tissues prior to hybridization that would affect the gut microbiota architecture.
In this report, we describe in detail the protocol to examine viable tick gut microbiota using whole-mount in situ hybridization (WMISH). The use of whole-mount RNA in situ hybridization enables a global understanding of the presence and abundance of specific gut bacteria in the different regions of the gut and may spur new insights into tick gut biology in the context of pathogen colonization and transmission. Further, the use of RNA probes directed against specific bacterial RNA allows detection of viable bacteria in the tick gut.
Protocol
1. Preparation of DNA Templates
- Download the 16S rRNA sequence specific to each bacterial genera from the NCBI database and design polymerase chain reaction (PCR) compatible forward and reverse primers that will amplify genera-specific regions (~200 – 250 base-pair long) as described earlier19. Also refer to the open source databases SILVA20,21 and probeBase22 online resources for rRNA-targeted oligonucleotide probes and primers, as RNA probes for some bacterial genera may be commercially available.
Note: It is important to select gene regions that are specific to specific genera and ensure that designed primers do not amplify related bacterial genera. This is critical to obtain gene/genera-specific probe hybridization. - Feed ticks for 24, 48 or 72 h on mice, dissect tick guts and prepare RNA and cDNA from tick guts as described earlier23. Use cDNA as template to PCR amplify genera-specific amplicons using a thermocycler. Run an aliquot of the PCR-product/amplicon on a 1.5% agarose gel and confirm that the amplicon corresponds to the expected size by visualization under ultraviolet (UV) light using a gel imaging system.
- Clone the bacterial 16S ribosomal RNA amplicon of interest into a commercially available TA vector (Table of Materials) containing RNA polymerase promoters suitable for bacteriophage polymerases (SP6, T7 or T3). Sequence the clones to confirm the identity of the amplicon and the directionality of the clone with respect to each of the promoters. Based on this, determine the promoter of choice to generate sense or antisense RNA probes (Figure 1).
- Linearize 1 mg of the plasmid with an appropriate restriction enzyme in a reaction volume of up to 30 µLfor 2 h at temperatures recommended by the manufacturer. Choose restriction enzymes based on the promoter that will be utilized to generate the sense or antisense probe (Figure 1). Verify linearization by visualization of 2 µL of digest reaction on a 1% agarose gel.
- Purify the remaining 28 µL of digested DNA using a commercially available PCR product column purification kit and quantify DNA concentration by spectrophotometer.
Note: Some sense probes may cause background staining much higher than the corresponding antisense probe and other options may need to be explored. Alternative negative controls include plasmids encoding sequences not represented in the sample or another probe that yields a distinctive pattern of hybridization.
2. Construction of Digoxygenin-UTP RNA Probes
- Prepare the nucleotide mix by combining 7.5 µL of 100 mM UTP, 25 µL of 10 mM digoxygenin-UTP, and 10 µL each of 100 mM ATP, GTP, and CTP in a 1.5 mL centrifuge tube.
- Perform in vitro transcription using commercially available T7 or Sp6 RNA polymerase kits, using 1 µL of the nucleotide mix (Step 2.1) and 0.25 – 1 µg of template DNA. Bring the volume up to 20 µL with water. Flick the tube to combine and pulse spin. Incubate at 37 °C for 2.5 – 3 h.
Note: Probe construction has been successful with digested plasmid concentrations as low as 7 ng/µL, but typical concentrations at this step range from 15 – 25 ng/µL. The transcription reaction may also be incubated overnight at 37 °C, but this does not significantly increase the RNA yield. - Add 1 µL of DNase (2,000 units/µL) and incubate at 37 °C for 10 min. Do not exceed 10 min, or the enzyme may begin degrading RNA.
- Add 30 µL of ice-cold 7.5 – 8 M lithium chloride and 30 µL of chilled nuclease-free water to precipitate the RNA. Flick the tube to mix. Allow precipitation of RNA to proceed overnight at -20 °C.
- Collect RNA by centrifugation at 17,000 x g for 20 min at 4 °C. Wash the pellet once with 1 mL of freshly prepared 75% ethanol in Diethyl pyrocarbonate (DEPC) water, then repeat centrifugation.
- Remove and discard the supernatant, invert the tube on a clean paper towel, and allow to dry for 10 min. Resuspend the pellet in nuclease-free water. If the pellet is easily visible, resuspend in 25 µL. If the pellet is very small or not visible, resuspend in 15 – 20 µL. Obtain an estimate of RNA concentration by spectrophotometer.
Note: Typical RNA concentrations range from 200 – 500 ng/µL.
3. Visualization of RNA Probes by RNA Formaldehyde Gel Electrophoresis to Assess RNA Purity
CAUTION: Formaldehyde poses an inhalation hazard; therefore, generate the solutions required for RNA gel analysis in a fume hood. Ethidium bromide is a suspected carcinogen; handle with care.
- Prepare a 1% formaldehyde agarose gel as previously described24 and cast the gel in a gel box free of RNases. Once cool, fill the gel box with 1x running buffer (1x MOPS at pH 7.0, 5% formaldehyde).
- Prepare the Sample Buffer (5 µL 400 mg/mL ethidium bromide, 20 µL 10x MOPS, 35 µL formaldehyde, and 100 µL formamide). Combine 2 µL of RNA probe (Step 2.6) with 8 µL of Sample Buffer. Incubate at 70 °C for 10 min.
- Prepare RNA Loading Buffer by combining 5 mL 50% glycerol, 1 mL 10% formaldehyde, 40 mg bromophenol blue, 40 mg xylene cyanol, and 20 µL of 0.5 M EDTA (pH 7.5). After use, store this buffer at -20 °C.
- Add 3 µL of Loading Buffer to the RNA sample from Step 3.2 and mix. Keep sample tubes on ice.
- Load the entire sample volume from Step 3.4 into the gel. Load an aliquot of single-stranded RNA ladder alongside to verify RNA size. Run the gel at 100 V or lower until the blue dye migrates approximately 75% of the way down the gel. Visualize the RNA under UV light using a gel imaging system.
Note: A good quality RNA probe should appear as a discrete band with a mobility corresponding to the expected size of the probe (Figure 2, lane 1). If the probe appears as a diffuse and smeary band (Figure 2, lane 2) this should not be used.
4. Tick Gut Collection and Fixation
- Collect Ixodes scapularis nymphs that have fed on mice for the time points of interest.
Note: For the purposes of this technique, ticks fed for 48 or 72 h work best. - Dissect the gut from the tick into MEMFA (Table 1) on a glass slide under a dissection microscope, avoiding damage to the organ as much as possible. Place a tick on a clean microscope slide in 20 – 30 µL of MEMFA. Using a pair of sterile high-carbon steel razor blade #11 slice the head region at the scutum of the tick leaving the body of the tick. Gently press the body to push the gut diverticula and salivary glands out the body cuticle. Separate the salivary glands and the gut and scoop the guts on the razor blade.
- Collect guts into a 1.5 mL tube containing 500 µL MEMFA. Incubate the tube at room temperature with gentle rocking for 1 h or overnight at 4 °C.
Note: Tick salivary glands may also be dissected and similarly fixed and stained. - Dehydrate the tissue by washing gently 3 times with 1 mL ice-cold 100% ethanol. Let the guts sink naturally to the bottom of the tube between each wash, as centrifugation may damage the tissue. For long-term storage, keep the samples in 100% ethanol at -20 °C.
5. Construction of Mesh Sample Baskets and 24-well Basket Holder
- Cut the bottoms off of 2 mL or 5 mL snap-top centrifuge tubes using a heated single-edged razor blade on a scalpel.
CAUTION: The blade may need to be reheated several times before the cut is completed. - Cut squares of a 110 µm nylon mesh slightly larger than the new tube opening. Bond the mesh to the bottom of the tube by pressing them together lightly onto a hot plate on medium-low heat until a good seal is made. Let cool, ensure there are no gaps between the mesh and the tube, and trim excess mesh around the edges.
- Cut holes in the cover of a 24-well plate such that the lip of the sample basket made in Step 5.2 will sit at the hole and not fall through, yielding a set-up where the bottoms of the sample baskets will sit all the way in the wells when they are placed in the holes in the cover.
- Use this fitted basket holder to transfer baskets from one plate to another with relative ease for the entirety of the in situ hybridization protocol. Use two 24-well plates so that while one incubation is occurring, the other plate is prepared for the next wash.
6. In Situ Hybridization: Day 1 (Timing: 4 – 5 h)
- Transfer guts to labeled sample baskets, separated by experimental condition and intended RNA probe.
Note: Stain at least 5 guts per condition to account for natural variation among replicates. - Rehydrate the samples gently to avoid introducing air bubbles into the tissue by gradually increasing the ratio of phosphate-buffered saline with 0.1 % non-ionic surfactant (PTw; Table 1) to ethanol in the washes.
Note: Complete all washes in the 24-well basket holder at ambient temperature with gentle agitation unless otherwise noted. Aliquot 500 – 600 µL of wash solutions into each well of the holder such that the guts in each basket are completely submerged. Prepare all solutions with DEPC-treated water to prevent RNA degradation.- Wash the samples for 5 min in 500 µL 100% ethanol.
- Prepare at least 500 µL per sample basket of 75%, 50%, and 25% ethanol in PTw. Rehydrate the samples by washing for 5 min in each ethanol solution, moving from the highest ethanol concentration to the lowest.
- Wash the samples 4 times for 5 min each in PTw.
- Permeabilize the samples with Proteinase K (10 µg/mL) in PTw for 5 min.
- Prepare the tissue for hybridization with the RNA probes.
- Wash the samples twice as described in 6.2 in the 24-well basket holder at ambient temperature with gentle agitation for 5 min each in 500 µL triethanolamine buffer (0.1 M, pH 7.0 – 8.0) (Table 1) to maintain the samples in an amine-free environment.
- Treat the samples twice with 500 µL 0.25% acetic anhydride in triethanolamine buffer (0.1 M, pH 7.0 – 8.0) for 5 min each, then wash the samples twice in PTw for 5 min each.
Note: The acetic anhydride acetylates free amines to neutralize positive charge, which is critical to prevent non-specific electrostatic interactions and ensure specific binding of the probe to mRNA. - Fix the guts for 20 min with 500 µL 4% paraformaldehyde in PTw, then wash 5 times in PTw for 5 min each.
- Prehybridize by incubating the samples in 500 µL hybridization buffer /in the 24-well basket holder in the 24 well plate (Table 1) for 1.5 – 2.5 h at 60 °C with gentle agitation in a shaking water bath. Save the hybridization buffer used for the prehybridization step to reuse in the Day 2 protocol (Step 7.1).
- Prepare probe hybridization solutions by diluting RNA probes in hybridization buffer to a final concentration of 1 µg/mL. Incubate samples with probe hybridization solutions in the 24-well basket holder at 60 °C with gentle agitation overnight in a shaking water bath (Table 2). Cover the plate carrying the basket holder snugly with aluminum foil to prevent evaporation of the hybridization solution.
7. In Situ Hybridization: Day 2 (Timing: 4.5 – 5.5 h)
- Remove the samples from the probe hybridization solutions and place them into the reserved hybridization buffer from the previous day. Incubate at 60 °C with gentle agitation for 3 min.
Note: Probe hybridization solutions may be stored at -20 °C for reuse up to 3 times. - Prepare 2x saline-sodium citrate buffer (SSC) by diluting 20x SSC in DEPC-treated water (Table 1). Wash the samples twice for 3 min with 2x SSC, then 3 times for 20 min with 2x SSC at 60 °C to remove all traces of probe and hybridization buffer.
- Treat with RNase A (20 µg/mL) and RNase T1 (10 µg/mL) in 2x SSC for 30 min at 37 °C to remove single stranded RNA probe representing free non-hybridized RNA.
- Wash once with 2x SSC for 10 min at room temperature. Prepare 0.2x SSC by diluting 2x SSC in DEPC-treated water, then wash twice with 0.2x SSC for 30 min at 60 °C to remove excess RNases.
- Wash twice with 500 µL of maleic acid buffer (MAB; Table 1) for 10 min each.
- Incubate samples with blocking solution (2% blocking reagent in MAB) for 1.5 – 2 h.
Note: The blocking reagent can be difficult to dissolve; gentle heating aids dissolution. - Replace the blocking solution with anti-digoxigenin alkaline phosphatase-conjugated 500 µL of antibody at a 1:3,000 dilution in blocking solution and incubate at room temperature for about 4 h or at 4 °C overnight.
8. In Situ Hybridization: Day 3 (Timing: 6 h to overnight)
- Wash the samples at least 5 times for 30 min each with 500 µL MAB to remove excess antibody.
Note: These washes are flexible and may be extended to 1 – 2 h, or 3 – 4 washes may be substituted for one overnight wash at 4 °C with minimal effect on efficacy. - Wash twice for 5 min with alkaline phosphatase buffer, pH 9.5 (AP buffer; Table 1).
- Begin the color reaction by replacing the last wash with 500 µL of the chromogenic substrate for alkaline phosphatase (Table of Materials). Cover the sample plate with aluminum foil and let the staining proceed at room temperature for 3- 5 h or overnight at 4 °C. Monitor the staining reaction periodically by viewing the tissue under a dissecting microscope to avoid overstaining.
Note: When the staining is complete, a purple tint is detectable by eye in the antisense-probed samples under the microscope, but the tissue should not be allowed to become very dark. Staining proceeds very slowly at 4 °C and may take up to 20 – 24 h. - Stop the color reaction by washing for 5 min in 500 µL MAB, then fix the tissue overnight in Bouin’s solution.
CAUTION: Handle the Bouin’s solution in a fume hood; it is a corrosive carcinogen.
9. In Situ Hybridization: Day 4 (Timing: 3 – 3.5 h)
- Prepare at least 7 mL per sample basket of 1x SSC by diluting 20x SSC in DEPC-treated water. Remove the Bouin’s solution by washing the samples 4 to 6 times with 70% ethanol in 1x SSC until the tissue is no longer yellow.
- Prepare 500 µL per sample basket of 75%, 50%, and 25% ethanol solutions in 1x SSC. Wash the samples for 5 min in each solution, moving from highest ethanol concentration to lowest to rehydrate the tissue. Wash twice for 5 min each in 1x SSC.
- Incubate the samples in the bleach solution (Table 1) for 90 min on a lightbox in a fume hood.
Note: This step allows any pigmentation in the samples or coloration from Bouins solution to be removed and enhances the probe signal for clear visualization. - Wash the samples twice with 500 µL of 1x SSC for 5 min.
- Relocate the guts with minimal damage by inverting the sample basket into a well of a new 24-well plate and wash with 70% ethanol through the mesh bottom using a transfer pipette. Store at -20 °C if needed.
- To obtain an overview of the staining patterns of multiple samples as shown in Figure 3, maintain the guts in 70% ethanol in the 24 well-plate and view with any bright-field microscope. To examine individual guts under a bright-field microscope as shown in Figure 4, Figure 5, and Figure 6, mount the guts in 100% glycerol under coverslips. Use a plastic spatula to gently manipulate the tissue with minimal damage.
Note: The high viscosity of glycerol helps protect the tissue from damage by compression and allows the careful positioning of diverticula such that the tissue can be viewed optimally at higher magnifications. - Capture images on the microscope using microscope specific image capture software (Table of Materials) and export as Tiff images to a generic image editing software. To quantify the relative staining differences between experimental treatments, download a generic image analysis software (Table of Materials). Open the image within the analysis software and use the instructions within the software to measure pixel intensity of the staining and compute histogram plots of the staining.
Representative Results
The measurement and estimation of the quality of the RNA probes are critical prior to beginning the staining. In vitro transcription efficiency depends highly on the amount and quality of the DNA template. We routinely visualized the RNA probes on a formaldehyde gel to verify the purity and amount of probe generated by the transcription reactions. The probes should appear as bright, discrete bands (Figure 2). Spectrophotometric measurements of RNA concentration were highly indicative of probe quality and correlated well with the appearance of the bands on the gel. Thus, the gel visualization step may be skipped if desired once familiarity with the protocol has been attained. Either way, it is important to ensure that sufficient RNA of high quality is produced, as all downstream steps depend upon the robustness of the probes.
The strength of this technique is in facilitating broad observations about the tick microbiota, including presence or absence of bacterial species, changes in abundance of bacteria over time as the tick feeds, and localization of species within the whole tissue. Some variation in staining amount and distribution is normal among biological replicates as well as among the 7 pairs of diverticula of individual guts (Figure 3), therefore, it is best to stain at least five guts per condition to get an idea of average staining pattern. The overall success of the protocol is best gauged by comparing the staining of the sense and antisense samples for each condition. The sense samples, important negative controls for this type of analysis, should have minimal to no purple color (Figure 4). Conversely, antisense samples should display robust staining with minimal background, the intensity of which will depend on the abundance of the specific bacterial transcripts being targeted. The staining pattern within the tissue will also vary based on these parameters. Importantly, the sense and antisense samples must be developed for the same amount of time in order to be able to directly compare them, so these samples should be processed in parallel. Overdevelopment of the color reaction can result in a high amount of background staining, where the sense samples begin to look similar to the antisense (Figure 5). Particular care should be taken at this step in the protocol to avoid overstaining, as there is no way to reverse the reaction once it occurs.
It is critical to make sure all guts are fully submerged in each reagent during every step; an increase in the variation across samples may be due to uneven exposure to reagents. The amount of time that the ticks are fed before the guts are collected also has a significant effect on the results. Ticks that were unfed or only fed for 24 h were difficult to work with; the guts were also more easily damaged and did not stain well (Figure 6) and is a limitation of this approach at this time. Ticks fed for 48 – 72 h were easiest to work with, due to the larger size of these guts when compared to guts from ticks fed for 24 h. Guts from later time points also stained better than 24 h-fed ticks, possibly due to increases in bacterial burden during the later stages of feeding. The 72-h tissue often had a slight background of brown tint from blood, but the purple staining was clearly visible over the tint (Figure 4). The guts are best viewed at low magnification (5X or 10X) to be able to view the staining patterns across the entire tissue using a bright-field microscope. Viewing whole-mount tissue at higher magnification, such as with a 63X oil objective, runs the risk of damaging the tissue as the objective would press against the slide and compress the tissue due to the thickness of the samples. If higher resolution imaging is desired, the potential for tissue sectioning should be examined.
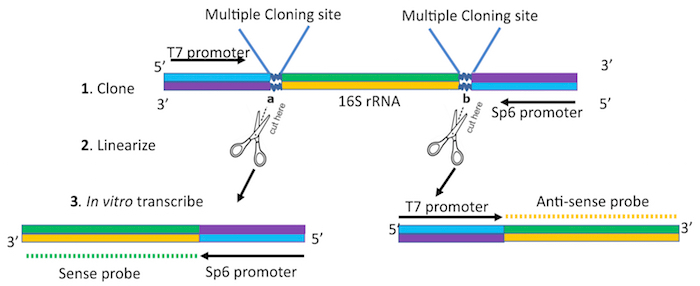
Figure 1. Schematic of template preparation for probe generation. Clone the 16S rRNA amplicon into a TA cloning vector containing T7 and Sp6 promoters. Linearize the construct using restriction enzymes to cut at sites a or b for in vitro transcription using T7 or Sp6 promoter to generate sense or antisense probes respectively. Please click here to view a larger version of this figure.
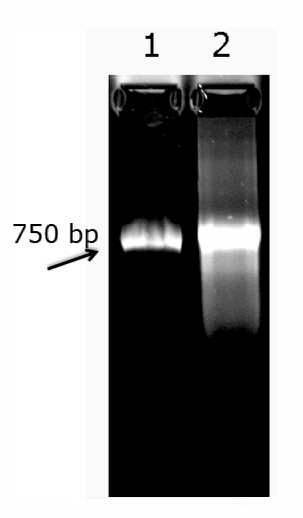
Figure 2. Visualization of the RNA probe by gel electrophoresis. RNA probes (~700 ng) were electrophoresed on a formaldehyde agarose gel, visualized under UV light, and imaged using a commercial gel imaging system. A representative good quality, lane 1; and poor-quality RNA probe, lane 2.
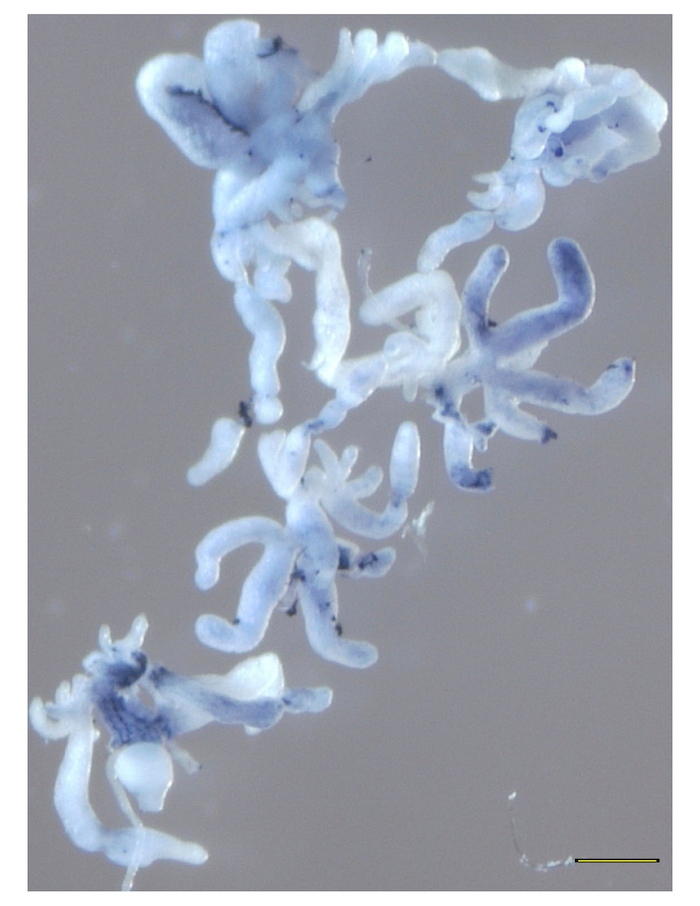
Figure 3. A representative overview of whole-mount in situ hybridization of 48 h-fed guts. Guts from ticks fed for 48 h stained with antisense RNA complementary to a conserved 16S ribosomal RNA sequence shows differential staining of gut diverticula. Scale bar represents 200 µm. Please click here to view a larger version of this figure.
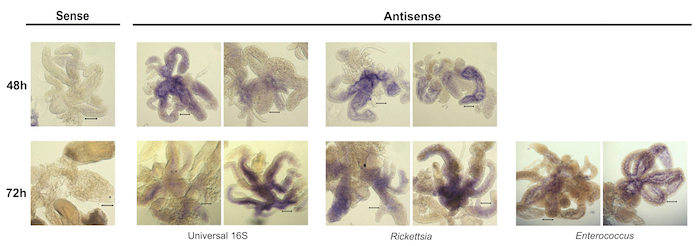
Figure 4. Representative images of successful whole-mount in situ hybridization in Ixodes scapularis guts. Guts from ticks fed for the indicated amount of time were stained using either sense (negative control) or antisense RNA probes. Antisense probes were complementary to a conserved 16S ribosomal RNA sequence (Universal 16S) or a 16S RNA sequence specific to a particular bacterial genus (as indicated). Enterococcus did not yield robust staining at 48 h. Scale bars represent 140 µm. Please click here to view a larger version of this figure.
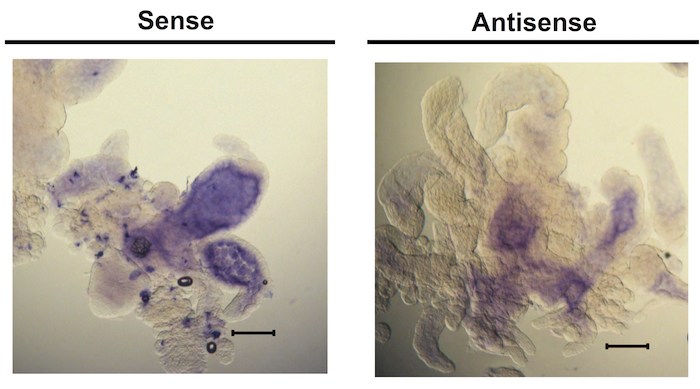
Figure 5. Prolonged chromogenic reaction time leads to non-specific staining. Guts from ticks fed for 48 h were probed with a sense RNA probe or an antisense probe complementary to a conserved 16S RNA sequence. The tissue was incubated with the alkaline phosphatase substrate BM purple overnight at 4 °C and then at room temperature for about 4 h before stopping the reaction. Scale bars represent 140 µm. Please click here to view a larger version of this figure.
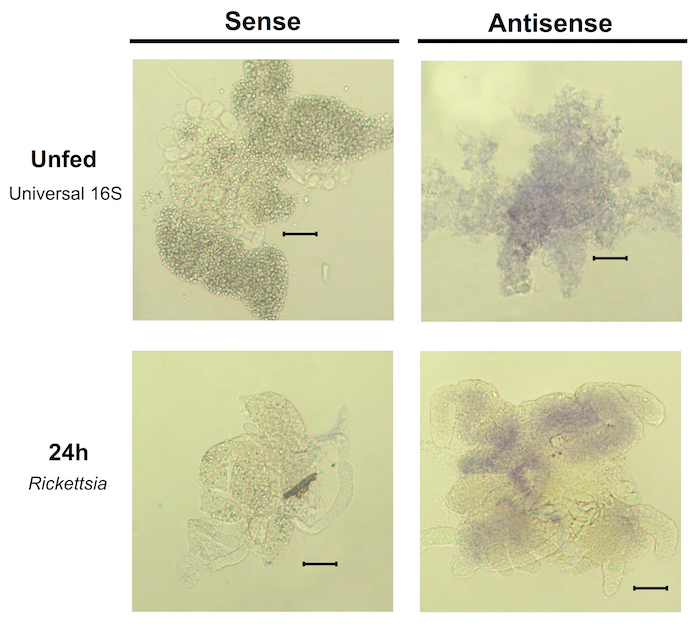
Figure 6. Whole-mount in situ hybridization using unfed or 24h fed tick guts. Guts from unfed or 24 h-fed ticks were stained using sense RNA probes or antisense probes complementary to a conserved 16S RNA sequence or a sequence specific to the genus Rickettsia. Staining in the antisense samples is less robust than observed at later time points and the guts are also more easily damaged. Scale bars represent 60 µm. Please click here to view a larger version of this figure.
| Name | Final Concentrations | Final pH | Storage notes |
| MEMFA | 0.1 M MOPS | – | Room temperature |
| 2 mM EGTA | |||
| 1 mM magnesium sulfate | |||
| 3.7% formaldehyde | |||
| PBS-Tween (PTw) | 1x PBS, 0.1% Tween-20 | 7.0 | Room temperature |
| 0.1% Tween-20 | |||
| Triethanolamine buffer | 1x PBS | 7.0 – 8.0 | Room temperature |
| 0.1 M triethanolamine | |||
| Hybridization buffer | 50% formamide | – | -20 °C |
| 5x SSC | |||
| 1 mg/mL Torula RNA | |||
| 100 µg/mL heparin | |||
| 1x Denhart's solution | |||
| 0.1% Tween-20 | |||
| 0.1% CHAPS | |||
| 10 mM EDTA | |||
| Maleic acid buffer (MAB) | 100 mM maleic acid | 7.0 | Room temperature |
| 150 mM sodium chloride | |||
| Alkaline phosphatase (AP) buffer | 100 mM Tris | 9.5 | -20 °C |
| 50 mM magnesium chloride | |||
| 100 mM sodium chloride | |||
| 0.1% Tween-20 | |||
| 5 mM levamisol | |||
| Bleach solution | 1% hydrogen peroxide | – | Prepare fresh |
| 5% formamide | |||
| 0.5x SSC |
Table 1. Recipes of solutions used in the protocol.
Discussion
This is the first use of a whole-mount in situ hybridization (WMISH) technique to study the microbiota of an arthropod vector of pathogens. Our protocol was adapted from one used to study development in Drosophila and in frog embryos25,26. Whole-mount RNA in situ hybridization has been routinely used to localize gene transcripts spatially and temporally27 and visualization of transcripts can be done by bright-field or by fluorescent microscopy. We have adopted the former towards detection of microbiota in tick guts. It is important to note that attempts to perform whole-mount in situ hybridization using whole ticks in which the head region was removed to allow penetration of fixative and hybridization solutions was not successful. The protocol described here utilizes dissected guts and has been implemented to study the effects of specific tick proteins on bacterial colonization in the midgut23. Information obtained from WMISH is comparable to immunohistochemistry but has the advantage of not requiring the generation of protein and antibodies to the gene or protein of interest. Genomic information is sufficient to generate the reagents for WMISH. Both WMISH and FISH (fluorescence in situ hybridization) can detect the gene expression. However, due to it being an enzymatic and chromogen-based assay, WMISH has the advantage that color development can be actively monitored, and the reaction is allowed to proceed until desired signal and sensitivity is achieved. It is fully amenable to automation and is easily scalable to high-throughput format. Unlike FISH and immunohistochemistry, no sectioning is required. The disadvantage of WMISH over FISH is that only 2 different mRNAs or genes can be addressed simultaneously and would require that probes be labeled with different nucleotide analogs such as digoxigenin-UTP and fluorescein-UTP28. With FISH, up to 6 different mRNAs can be examined in one assay29. While both assays require comparable time, the cost of fluorescence dyes are higher than that of chromogenic substrates. FISH assays would also require a fluorescence microscope for image visualization, while WMISH image visualization can be performed with any light microscope.
It is crucial that the protocol is followed closely; however, there are some steps that require particular care. Dissection of the midgut from the tick without damaging the tissue requires some dexterity. It may be prudent to collect extra ticks to practice the dissections and gain proficiency. Due to the heterogeneity of staining in the different diverticula of individual guts (Figure 3), it is important to examine all the diverticula of each gut to derive conclusive evidence of bacterial abundance. It is also important to process several guts (at least 5) for each experimental condition in order to obtain conclusive information on the presence, spatial distribution, and abundance of specific bacteria within different diverticula. Production of high-quality RNA probes is also critical to effective tissue staining. It is crucial to also confirm the sequence of the probe template in the cloning vector to ensure that antisense and sense probes are appropriately generated. A productive in vitro transcription reaction requires sufficient template DNA; a minimum of 100 ng may be used, but 0.5 – 1 µg is recommended for optimal probe generation.
The use of sample baskets and 24-well plates in this protocol avoids direct manipulation of the tissue at every wash step, which minimizes damage during the staining process. However, samples are still occasionally lost or damaged; guts from unfed or 24h-fed ticks, in particular, are difficult to see and easy to lose when transferring from the sample baskets to a long-term storage container. Additional care should be taken while handling these samples. Due to this limitation, we have mainly used 48 and 72 h-fed tick guts. The presence of large amounts of blood meal in the guts at these time-points (longer than 72 h) interferes with the staining and visualization due to non-specific background from the blood-meal and also presents a limitation of this technique. It is likely that the use of fluorescently-labeled probes might circumvent this issue.
The most important aspect of the staining procedure is to establish the balance between robust staining and low background signal. The timing of ideal color development can vary depending on the experimental conditions. Thus, it is best to perform the color reaction at room temperature and monitor color development approximately every 30 min. The reaction may also be set up to proceed overnight at 4 °C to slow color development. However, it is extremely critical not to let this reaction proceed for long. The staining reaction can be difficult to monitor by eye, so the samples should be examined under a dissection microscope. A purple tint should be visible on samples probed with antisense RNA, while the samples probed with sense RNA should retain minimal color. If the sense RNA-probed samples stain brightly, troubleshooting steps should begin with reducing the incubation time with BM purple and increasing the blocking time. Sometimes sense probes can cause anomalously high background compared to the antisense probes; in these cases, different regions of the gene might have to be considered for generating RNA probes. DNA sequences not represented in a given sample may also be considered as templates to generate negative control probes. For example, a DNA template encoding a region of the green fluorescent protein, not represented in the tick genome or its microbiome, can be utilized.
One limitation of this technique is that the analysis is largely qualitative; however, image analysis software (Table of Materials) could be used to measure the amount of staining signal in each sample. This would allow a semi-quantitative comparison of the abundance of various bacterial species under different experimental conditions. While this is a time-consuming protocol that takes 3 – 4 days to complete, it is not laborious; each day hands-on time is only about 3 – 5 h on average. However, the ability to process many samples in parallel with the use of the sample baskets and holders allows for high-throughput analysis. Further, this process can be automated using commercially available instruments (Table of Materials) if this protocol is expected to be an essential and routine component of the research laboratory by using commercially available automation-compatible platforms (Table of Materials) to further reduce hands-on time.
The described protocol makes use of digoxygenin-labeled probes that allow the production of a colorimetric signal that can be imaged using bright field microscopy. While we have utilized one chromogenic substrate to detect hybridized RNA probes specific to individual targets, it is also possible to detect multiple targets simultaneously by labeling probes with different nucleotide analogs, such as digoxigenin-UTP and fluorescein-UTP and different chromogenic substrates30. The tissue samples can then be incubated sequentially with alkaline-phosphatase-coupled antibodies against digoxigenin and fluorescein, with different chromogenic reactions for the two steps. This technique could also be adapted for fluorescently-labeled probes, which may facilitate higher-resolution imaging using confocal microscopy31 and provide the option of using multiple probes of different colors in parallel. This technique can only specifically assess a few microorganisms at one time in one sample. However, several samples can be assessed in high-throughput assays to address multiple microorganisms that may be represented in tick microbiota.
Finally, this technique is not limited to the investigation of the interactions between ticks and their associated microbiota. Probes complementary to any gene of interest within the tick genome may be generated and used to examine the abundance and localization of specific genes in different tissues. The salivary glands of the tick can also be processed and analyzed similarly to the gut. Overall, this technique has broad potential for adaptation to studies on the dynamics of microbial communities within other arthropod disease vectors of interest.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We sincerely thank Dr. Mustafa Khokha, Yale University, for providing the use of his laboratory resources. We are grateful to Mr. Ming-Jie Wu for excellent technical assistance. EF is an HHMI investigator.This work was supported by a gift from the John Monsky and Jennifer Weis Monsky Lyme Disease Research Fund.
Materials
| Sefar NITEX Nylon Mesh, 110 micron | Amazon | 03-110/47 | |
| pGEM-T Easy Vector System | Promega | A1360 | |
| Digoxygenin-11-UTP | Roche | 1209256910 | |
| dNTP | New England Biolabs | N0447S | |
| DNAse I(RNAse-free) | New England Biolabs | M0303S | |
| HiScribe SP6 RNA synthesis kit | New England Biolabs | E2070S | |
| HiScribe T7 High Yield RNA Synthesis Kit | New England Biolabs | E2040S | |
| Water, RNase-free, DEPC-treated | American Bioanalytical | AB02128-00500 | |
| EDTA, 0.5M, pH 8.0 | American Bioanalytical | AB00502-01000 | |
| Formaldehyde, 37% | JT Baker | 2106-01 | |
| Formamide | American Bioanalytical | AB00600-00500 | |
| EGTA | Sigma Aldrich | E-4378 | |
| DPBS, 10X | Gibco | 14300-075 | |
| Tween-20 | Sigma Aldrich | P1379-25ML | |
| Proteinase K | Sigma Aldrich | 3115879001 | |
| Triethanolamine HCl | Sigma Aldrich | T1502-100G | |
| Acetic anhydride | Sigma Aldrich | 320102-100ML | |
| Paraformaldehyde | ThermoScientific/Pierce | 28906 | |
| SSC, 20X | American Bioanalytical | AB13156-01000 | |
| RNA from torula yeast | Sigma Aldrich | R3629-5G | |
| Heparin, sodium salt | Sigma Aldrich | H3393-10KU | |
| Denhardt's Solution, 50X | Sigma Aldrich | D2532-5ML | |
| CHAPS hydrate | Sigma Aldrich | C3023-1G | |
| RNase A | Sigma Aldrich | 10109142001 | |
| RNase T1 | ThermoScientific | EN0541 | |
| Maleic acid | Sigma Aldrich | M0375-100G | |
| Blocking reagent | Sigma Aldrich | 11096176001 | |
| Anti-Digoxigenin-AP, Fab fragments | Sigma Aldrich | 11093274910 | |
| Levamisol hydrochloride | Sigma Aldrich | 31742-250MG | |
| Chromogenic substrate for alkaline phosphatase | Sigma Aldrich | 11442074001 | |
| Bouin's solution | Sigma Aldrich | HT10132-1L | |
| Hydrogen peroxide | Mallinkrodt Baker, Inc | 2186-01 | |
| Single stranded RNA ladder | Ambion -Millenium | AM7151 | |
| #11 High-Carbon steel blades | C and A Scientific Premiere | #11-9411 | |
| Thermocycler | BioRad, CA | 1851148 | |
| Spectrophotometer | ThermoScientific | NanoDrop 2000C | |
| Orbital shaker | VWR | DS-500E Digital Orbital shaker | |
| Shaking water bath | BELLCO Glass, Inc | Hot Shaker-7746-12110 | |
| Gel documentation system | BioRad | Gel Doc XR+ Gel documentation system | |
| Bright-field Microscope | Nikon | NikonSM2745T | |
| Bright-field Microscope | Zeiss | AXIO Scope.A1 | |
| Dissection microscope | Zeiss | STEMI 2000-C | |
| Light box | VWR | 102097-658 | |
| PCR purification kit | Qiagen | 28104 | |
| Image capture software | Zeiss | Zen lite | |
| Image editing software | Adobe | Adobe Photoshop CS4 version 11.0 | |
| Image analysis software | National Institutes of Health | ImageJ-NIH /imagej.nih.gov/ij/ | |
| Automation compatible instrumentation | Intavis Bioanalytical Instruments, Tubingen, Germany). | Intavis, Biolane HT1.16v |
References
- Leitner, W. W., Wali, T., Kincaid, R., Costero-Saint Denis, A. Arthropod Vectors and Disease Transmission: Translational Aspects. PLoS Neglected Tropical Diseases. 9 (11), e0004107 (2015).
- Hooper, L. V., Gordon, J. I. Commensal host-bacterial relationships in the gut. Science. 292 (5519), 1115-1118 (2001).
- Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R., Gordon, J. I. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Review Microbiology. 6 (10), 776-788 (2008).
- Narasimhan, S., Fikrig, E. Tick microbiome: the force within. Trends in Parasitology. 31 (7), 315-323 (2015).
- Weiss, B., Aksoy, S. Microbiome influences on insect host vector competence. Trends in Parasitoly. 27 (11), 514-522 (2011).
- Degli Esposti, M., Martinez Romero, E. The functional microbiome of arthropods. PLoS One. 12 (5), e0176573 (2017).
- Radolf, J. D., Caimano, M. J., Stevenson, B., Hu, L. T. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nature Reviews Microbiology. 10 (2), 87-99 (2012).
- Sojka, D., et al. New insights into the machinery of blood digestion by ticks. Trends in Parasitology. 29 (6), 276-285 (2013).
- Grigor’eva, L. A. Morphofunctional changes in the midgut of Ixodid ticks during the life cycle. Entomological Review. 90, 405-409 (2010).
- Goodrich, J. K., et al. Conducting a microbiome study. Cell. 158 (2), 250-262 (2014).
- Salter, S. J., et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biology. 12, 87 (2014).
- Hemmati-Brivanlou, A., et al. Localization of specific mRNAs in Xenopus embryos by whole-mount in situ hybridization. Development. 110 (2), 325-330 (1990).
- Frank, D., Harland, R. M. Transient expression of XMyoD in non-somitic mesoderm of Xenopus gastrulae. Development. 113 (4), 1387-1393 (1991).
- Harland, R. M. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods in Cell Biology. 36, 685-695 (1991).
- Kernohan, K. D., Berube, N. G. Three dimensional dual labelled DNA fluorescent in situ hybridization analysis in fixed tissue sections. MethodsX. 1, 30-35 (2014).
- Sonenshine, D. E. . Biology of ticks. , (1991).
- Narasimhan, S., et al. Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the lyme disease spirochete. Cell Host Microbe. 15 (1), 58-71 (2014).
- Hammer, B., Moter, A., Kahl, O., Alberti, G., Gobel, U. B. Visualization of Borrelia burgdorferi sensu lato by fluorescence in situ hybridization (FISH) on whole-body sections of Ixodes ricinus ticks and gerbil skin biopsies. Microbiology. 147 (Pt 6), 1425-1436 (2001).
- Harmsen, H. J., Raangs, G. C., He, T., Degener, J. E., Welling, G. W. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Applied Environmental Microbiology. 68 (6), 2982-2990 (2002).
- Quast, C., et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research. 41 (Database issue), D590-D596 (2013).
- Yilmaz, P., et al. The SILVA and "All-species Living Tree Project (LTP)" taxonomic frameworks. Nucleic Acids Research. 42, D643-D648 (2014).
- Greuter, D., Loy, A., Horn, M., Rattei, T. probeBase–an online resource for rRNA-targeted oligonucleotide probes and primers: new features 2016. Nucleic Acids Research. 44, D586-D589 (2016).
- Narasimhan, S., et al. Modulation of the tick gut milieu by a secreted tick protein favors Borrelia burgdorferi colonization. Nature Communication. 8 (1), 184 (2017).
- Bryant, S., Manning, D. L. Formaldehyde gel electrophoresis of total RNA. Methods Molecular Bioliogy. 86, 69-72 (1998).
- Tautz, D., Pfeifle, C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 98 (2), 81-85 (1989).
- Robson, A., Owens, N. D., Baserga, S. J., Khokha, M. K., Griffin, J. N. Expression of ribosomopathy genes during Xenopus tropicalis embryogenesis. BMC Development Biology. 16 (1), 38 (2016).
- Khokha, M. K., et al. Techniques and probes for the study of Xenopus tropicalis development. Developmental Dynamics. 225 (4), 499-510 (2002).
- Jowett, T., Lettice, L. Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin- and fluorescein-labelled probes. Trends in Genetics. 10 (3), 73-74 (1994).
- Behnam, F., Vilcinskas, A., Wagner, M., Stoecker, K. A straightforward DOPE (double labeling of oligonucleotide probes)-FISH (fluorescence in situ hybridization) method for simultaneous multicolor detection of six microbial populations. Applied Environmental Microbiology. 78 (15), 5138-5142 (2012).
- Pai, V. P., et al. HCN4 ion channel function is required for early events that regulate anatomical left-right patterning in a nodal and lefty asymmetric gene expression-independent manner. Biology Open. 6 (10), 1445-1457 (2017).
- Legendre, F., et al. Whole mount RNA fluorescent in situ hybridization of Drosophila embryos. JoVE. (71), e50057 (2013).

