High Throughput Traction Force Microscopy Using PDMS Reveals Dose-Dependent Effects of Transforming Growth Factor-β on the Epithelial-to-Mesenchymal Transition
Summary
We present a high throughput traction force assay fabricated with silicone rubber (PDMS). This novel assay is suitable for studying physical changes in cell contractility during various biological and biomedical processes and diseases. We demonstrate this method’s utility by measuring a TGF-β dependent increase in contractility during the epithelial-to-mesenchymal transition.
Abstract
Cellular contractility is essential in diverse aspects of biology, driving processes that range from motility and division, to tissue contraction and mechanical stability, and represents a core element of multi-cellular animal life. In adherent cells, acto-myosin contraction is seen in traction forces that cells exert on their substrate. Dysregulation of cellular contractility appears in a myriad of pathologies, making contractility a promising target in diverse diagnostic approaches using biophysics as a metric. Moreover, novel therapeutic strategies can be based on correcting the apparent malfunction of cell contractility. These applications, however, require direct quantification of these forces.
We have developed silicone elastomer-based traction force microscopy (TFM) in a parallelized multi-well format. Our use of a silicone rubber, specifically polydimethylsiloxane (PDMS), rather than the commonly employed hydrogel polyacrylamide (PAA) enables us to make robust and inert substrates with indefinite shelf-lives requiring no specialized storage conditions. Unlike pillar-PDMS based approaches that have a modulus in the GPa range, the PDMS used here is very compliant, ranging from approximately 0.4 kPa to 100 kPa. We create a high-throughput platform for TFM by partitioning these large monolithic substrates spatially into biochemically independent wells, creating a multi-well platform for traction force screening that is compatible with existing multi-well systems.
In this manuscript, we use this multi-well traction force system to examine the Epithelial to Mesenchymal Transition (EMT); we induce EMT in NMuMG cells by exposing them to TGF-β, and to quantify the biophysical changes during EMT. We measure the contractility as a function of concentration and duration of TGF-β exposure. Our findings here demonstrate the utility of parallelized TFM in the context of disease biophysics.
Introduction
Acto-myosin contractility is an essential element of active cell mechanics, impacting cell behaviors from motility and proliferation to stem cell differentiation. In tissues, contractility drives activity from polar separation in embryogenesis, to airway constriction and cardiac activity. Critically, to generate tension, cells must first adhere to their extracellular environment. In doing so, this contractility generates traction forces on their surroundings. Traction Force Microscopy (TFM) has emerged in a multitude of forms as a way to quantify these forces from diverse cells under different conditions.
The field of TFM has seen an exceptional breadth of innovation and application, and the results have paved the way for new perspectives in biology, which incorporate mechanics and physical forces. Starting with wrinkling silicone substrates1, researchers have applied various techniques to measure cell traction forces. These approaches have been continuously improved and have now reached a level of resolution on the order of several microns2. However, one principal problem has emerged, which is the difficulty in creating substrates of suitably low moduli using the available silicones. To circumvent this problem, polyacrylamide was adopted as a replacement due to the ease of creating substrates on the order of 1-20 kPa3. We recently implemented very compliant silicones in TFM4, allowing us to fabricate the same range of moduli as polyacrylamide, but with the advantages of inert and robust silicone.
TFM approaches have enabled valuable mechano-biological discoveries, however, a persistent shortcoming is their complexity, often restricting their use to researchers in the engineering or physical sciences disciplines. This is due in large part to the detailed calibrations and challenging calculations that are required to quantify contractility. Another significant challenge is that TFM methods are largely low-throughput and therefore ill-suited to study many different conditions or populations simultaneously5. This has presented a bottleneck, which has hampered transfer of TFM from a specialist biophysics setting into broader biological and pharmacological applications.
We have recently developed a multi-well format TFM plate, which allows researchers to parallelize their TFM measurements for faster quantification of contractility metrics, while exploring the impact of different compounds and also using less reagents4. This methodology has broad utility in diverse mechanobiology studies, from evaluating the effects of compounds on cellular activity, to quantifying the contractile changes in differentiation or disease.
One area of biomedical research that will benefit greatly from TFM is the study of how physical cues impact the malignant phenotypes of cancer cells. Metastasis, responsible for 90% of cancer-related deaths, is characterized by cancerous cells leaving their original tumor site and colonizing a secondary site. For cells to migrate through tissue and pass in and out of the vascular system, they must radically change their shapes to squeeze through these physical barriers while generating substantial forces to pull their way along extracellular matrix or move between other cells. These forces are transmitted to the substrate through focal adhesion interactions2,3, and can be quantified using TFM. While cancers are biochemically exceptionally diverse, with an expanding repertoire of known mutations and protein changes, some common physical changes have been observed; in a variety of cancers, including breast, prostate, and lung cancers, metastatic cells have been shown to exert 2-3 times the traction forces of non-metastatic cells6,7,8. These results suggest that there may be a strong correlation between metastatic progression and the traction forces exerted by cells; however, the detailed time-dependent changes in contractility are difficult to examine.
The epithelial-to-mesenchymal transition (EMT) is a process whereby cells reduce adherens- and tight-junction mediated cell-cell adhesion, becoming more migratory and invasive. In addition to physiological functions that include wound healing and developmental processes, EMT is also a process exploited during metastasis, making it a useful model system to study this process. Using TGF-β, we can induce the EMT in murine mammary epithelial cells (NMuMG)9 to directly quantify the physical changes during this transformation, and characterize the time and dose-dependent effects of TGF-β on EMT and cell contractility. In this article, we demonstrate the utility of this approach by measuring the changes in contractility during an induced EMT.
Protocol
NOTE: The following protocol will guide researchers in fabricating and using the multi-well TFM dish shown in Figure 1.
1. Preparation of PDMS silicone substrates
- Preparation of PDMS silicone rubber mixture based on a composite mixture of two commercially available kits.
- Add Part A and Part B of PDMS kit (e.g., GEL-8100, see the Table of Materials) in a 1:1 weight ratio into the 50 mL tube.
NOTE: The mixture is mixed on a rotator at a speed slow enough for the mixture to flow back and forth during revolution to ensure complete mixing. - Add the required amount of curing agent for the desired modulus of the substrate.
NOTE: The amount of curing agent to be added to the mixture depends on the desired modulus of the substrate and may typically range from 0.1% to 1.8%. Refer to Table 1 and Figure 3 for a guide to specific crosslinker concentrations and resulting moduli. - Mix the formulation on the rotator for 30-45 min; ensure the rotation is slow enough for thorough mixing.
- Add Part A and Part B of PDMS kit (e.g., GEL-8100, see the Table of Materials) in a 1:1 weight ratio into the 50 mL tube.
- Bottom layer: coating PDMS substrates on the glass slide
- Place the custom-built chuck illustrated in Figure 2 on the spin-coater. Clean the glass with ethanol or isopropanol, and dry with a lint-free wipe. Place the glass slide in the chuck and turn on the vacuum to hold the slide in place.
- Apply uncured PDMS approximately 1 cm from the edges and work in towards the center. Apply enough (3-4 mL) PDMS to ensure the whole surface will be covered.
NOTE: To ensure that the PDMS is evenly spread on the surface of the glass, a pipette tip may be helpful to spread the PDMS mixture from the center to the edges. - Spin the glass with the PDMS mixture on a spin-coater with the following protocol.
- To spread the uncured PDMS on the slide, accelerate at 50 rpm/s from 0 to 200 rpm; hold at 200 rpm for 1 min.
- To achieve a 100 µm thick PDMS layer, accelerate at 50 rpm/s to 300 rpm and hold for 1 min at 300 rpm. Different desired thicknesses other than 100 µm will require specific speed values.
- To remove, decelerate at 50 rpm/s to 0 rpm. Disable vacuum and remove the coated slide, taking care not to touch the coated surface.
NOTE: It is important to include the acceleration and deceleration steps to ensure a smooth continuous surface.
CAUTION: To ensure that the sample does not fly off the chuck, the custom-made holder should be used to hold the slide, and not rely simply on vacuum and the existing flat chuck. The details and specifications of this holder are given in Figure 2.
- Place the spin-coated sample in the oven at the manufacturer recommended temperature (100 °C) for 2 h.
NOTE: The surface of the oven where the sample is placed should be solid (i.e., not a wire rack) and level surface to ensure the uniform heating and thickness of the sample. A ceramic or steel plate makes an ideal surface.
- Top bead layer
- Add bead solution in the appropriate ratio to the remaining PDMS mixture.
NOTE: This ratio depends on the concentration of the stock bead solution and the desired bead density on the sample. Typical final values are 9.2 x 1011 beads/mL and 0.05-0.2 beads/µm2, and an excess of beads is generally preferable to an inadequate amount. - Mix the bead solution with the uncured PDMS. This may be accomplished by placing the tube on a rotator for approximately 30 min, vortexing for 1-2 min, or sonication for 30 min. These methods may be combined.
NOTE: In our application, we find that sonication is effective in breaking bead aggregates, and rotation is effective in mixing. Synthesized fiduciary beads may aggregate in storage. Prior to use, they may be resuspended in hexane and sonicated. If there are significant large aggregates, one can filter the bead suspension through a 5 µm syringe filter. This filtration step is optional; it helps to coat the sample with monodispersed beads, but a significant fraction of beads may be lost in the filter. - Take out the slide from the oven, allow to cool to room temperature, and place it on the spin-coater.
- Add 3-4 mL of the bead and uncured PDMS mixture onto the surface of the coated sample.
NOTE: The mixture with beads added is less viscous due to the hexane. Make sure not to touch the surface of the substrate as it may damage the already-coated PDMS substrate. Additionally, the bead mixture may initially not wet the surface; take care that the mixture does not immediately flow off the cured PDMS surface. - Spin the sample with the following protocol.
- To spread the bead and uncured PDMS mixture, accelerate at 100 rpm/s from 0 to 500 rpm; hold at 500 rpm for 1 min.
- To achieve a thin layer of bead-embedded PDMS (~1 µm), accelerate at 200 rpm/s from 500 to 5000 rpm; hold at 5000 rpm for 10 s.
- To remove, decelerate at 100 rpm/s to 0 rpm. Disable vacuum and remove coated slide, taking care not to touch the coated surface.
- Place the spin-coated sample in the oven at 100 °C for 1 h.
NOTE: Temperatures above 100 °C or durations longer than 1 h can reduce the bead fluorescence. Make sure that the oven temperature is set to 100 °C and not higher. - The protocol can be paused here. To store the sample, cover the surface to avoid dust and light exposure. Make sure that nothing touches the surface. The sample is shelf-stable at room temperature indefinitely.
- Add bead solution in the appropriate ratio to the remaining PDMS mixture.
- Assembling the plate
- Add Part A (base) and Part B (curing agent, e.g., Sylgard) of a PDMS elastomer kit in a 10:1 weight ratio into the 50 mL tube.
- Mix the mixture on the rotator for 30-45 min.
NOTE: For one plate, mix 5 mL of base with 0.5 mL of curing agent. Up to 1 mL of hexane can be added to reduce the viscosity of the mixture. - Apply the mixture to the bottom of the divider and spread the mixture.
NOTE: The divider should be placed upside-down. - Lay the substrate onto the divider upside-down.
- Place the sample upside-down in the oven at 65 °C for 2 h.
- Take out samples from the oven and clean the bottom of the glass with 70% ethanol or isopropanol to remove any PDMS residue.
NOTE: The protocol can be paused here. To store the sample, place a lid on the substrate and wrap the device in aluminum foil to avoid exposure to the light. The sample is shelf-stable at room temperature indefinitely. The divider utilized in this method is in a 96-well format; however, researchers may employ other formats (384-well, 2-well, 4-well, 8-well, etc.) depending on desired experiment setups and availability of dividing structures. Some further optimization may be required.
2. Surface functionalization
- Dissolve 80 µL of a Sulfo-SANPAH aliquot in 40 mL of 0.1 M HEPES buffer (pH 7-9).
NOTE: Prepare Sulfo-SANPAH aliquots by dissolving 100 mg of Sulfo-SANPAH powder in 2 mL of sterile dimethyl sulfoxide (DMSO). Prepare 0.1 M HEPES buffer by diluting 50 mL of HEPES in 450 mL of sterile deionized water and filter through a 0.22 µm pore filter. - Add 200 µL of diluted Sulfo-SANPAH solution to each well of the 96-well plate.
- Expose the plate to UV (300-460nm) light at appropriate distance and duration.
NOTE: After UV exposure, the color of the solution should be darker. UV exposure distance and duration depend on the UV lamp power. In our application, we expose for 10-15 min at a distance of 5 cm. - Remove the Sulfo-SANPAH solution from the wells and add 200 µL of 5 µg/mL fibronectin solution to each well.
NOTE: Researcher-specified protein can be used for surface coating. Some commonly used proteins are collagen, fibronectin, and laminin. We have found Sulfo-SANPAH to be the most effective method, and plasma cleaning while sometimes employing in PDMS is discouraged as it creates a silicon dioxide layer and visibly damages the surface. - Incubate the plate at 4 °C overnight.
NOTE: Different incubation methods can be applied depending on the protein used for coating. - Remove the fibronectin solution and wash each well with phosphate-buffered saline (PBS) twice.
- Place a lid on the sample.
- Add 200 µL of PBS to each well.
NOTE: The protocol can be paused here. The fibronectin-coated samples can be stored at 4 °C for up to 2 weeks.
3. UV sterilization
- Sterilize the sample under UV light in a biological safety cabinet for 30 min.
NOTE: Longer UV exposure times may negatively impact the bead fluorescence. All subsequent steps must be performed under sterile conditions.
4. Cell culture
- Remove PBS from each well and add 200 µL of cells suspended in culture media to each well.
- Plate the cells at the desired cell density. Cell density depends on the desired experiment. For single cell studies, cells should be minimum of 50 µm apart and cells near the edges of the imaging window should be not be included in the TFM measurement. For monolayer cells, the imaging window should have the viewing field covered with a confluent layer of cells.
- Prepare the complete growth culture media for NMuMG cells by supplementing DMEM with 5% FBS, 10 mM HEPES, 10 µg/mL insulin, 1% penicillin-streptomycin, 1 mM L-glutamine, and 0.5 µg/mL amphotericin B.
- Prepare insulin stock by reconstituting in acidified water (2.5 mL of glacial acetic acid in 130 mL of deionized water) to a concentration of 10 mg/mL. Store the stock solution at 4 °C. Wait until the solution is clear, and then filter through a 0.22 µm pore filter.
- TGF-β addition
- To prepare of TGF-β stock, dissolve 2 µg of TGF-β in 100 µL of 10 mM citric acid (pH 3.0) and filter sterilize with 0.22 µm pores. Vortex the tube and aliquot into the desired volumes. Store the aliquots at -80 °C.
- Add 1.5 µL of TGF-β stock solution to 10 mL of the complete cell culture media to constitute the cell culture media with the final TGF-β concentration of 3 ng/mL.
NOTE: To make 10 mM pH 3.0 citric acid, dilute the acid in water and adjust pH to 3.0 by adding HCl.
5. Data acquisition
- For each position, acquire at least one image of fiduciary particles and cells. Focus on the bead layer.
NOTE: Pixel size should be optimized based on size of the fiduciary particles and image processing method being used. In this application, the authors use a 10x 0.4 NA objective, and images are acquired with 1024 x 1024 resolution, with 455 nm/pixel. In general, it is helpful to retain a resolution of at least approximately 1-5 pixels per bead; here, beads are polydisperse and have an individual size of 300-500 nm. It is critical that the fluorescent fiduciary beads be in focus for images to be used for TFM calculations. The focus and imaging quality of the beads should be prioritized over imaging the cells themselves. There should be no cross-talk between different channels, particularly any fluorescence not from the fiduciary beads which appears in the imaging spectra of the beads. - Once all the positions of interest have been recorded, add detachment solution to each well to acquire force-free reference images of the fiduciary particles.
- To prepare cell detachment solution, mix an aqueous solution containing 2% TritonX-100, 50 mM sodium azide, and 500 mM potassium hydroxide.
NOTE: The above is provided as an example of an effective detachment solution. Different detachment solutions at researchers' discretion may be used to detach the cells off the substrate surface.
- To prepare cell detachment solution, mix an aqueous solution containing 2% TritonX-100, 50 mM sodium azide, and 500 mM potassium hydroxide.
6. Image analysis
- Perform appropriate image analysis as desired.
NOTE: Analysis software was developed in-house. Image analysis may be done with custom-made software or software available online.
7. Bead synthesis
NOTE: The following protocol is based on the synthesis method described by Klein et al.10.
- Under a fume hood, prepare the three-neck flask with a water-cooled reflux condenser.
CAUTION: Set up a synthesis in a well-ventilated chemical fume hood. - Add 0.5 mL of PDMS stabilizer and fluorophore to the flask.
- Equip one neck with a rubber septum with a nitrogen inlet needle and an outlet needle and equip the other neck with a rubber septum for adding reagent with a syringe.
- Add 100 mL of anhydrous hexane in 250 mL to the flask and add a small magnetic stir bar.
- Place the flask in the mineral oil bath at 75 °C and purge it with nitrogen gas for 1 h.
- Add 6 mL of methylmethacrylate to a 25 mL round bottom flask.
- Add 0.100 g of 2,2'-azobisisobutyronitrile (AIBN) to the round bottom flask and purge the mixture with nitrogen gas for 1 h.
NOTE: Flush methylamethacrylate through prepacked column to remove inhibitors before use. Add methylamethacrylate and AIBN mixture to the three-neck flask. - The solution initially becomes cloudy and turns milky. Let the reaction run for 3 h after the solution becomes cloudy.
- After 3 h, place the flask in an ice water bath.
- Vacuum-filter the solution with coarse filter paper.
- Centrifuge the filtrate and re-suspend the particles in hexane.
NOTE: The volume of the hexane to be added depends on the desired concentration of the bead solution. Sonication facilitates re-dispersion of the bead particles in hexane solution. Beads produced by the authors have polydisperse diameters of approximately 300-500nm. Due to the use of cross-correlation pattern tracking to determine displacements, monodisperse beads are not required.
8. Rheology measurement protocol
NOTE: Rheology is not required for every researcher or experiment, but is necessary to quantify the moduli for new formulations of PDMS. In this protocol, we employ a shear rheometer to measure the effects of crosslinker, frequency, and strain on moduli of PDMS samples. Depending on the available tools and expertise, moduli may also be measured using many other mechanical analysis approaches. Additionally, researchers using this protocol may elect to use our published moduli presented in Table 1, Figure 3 and Figure 4.
- Use a rheometer with a 25 mm diameter parallel plate geometry. Other geometries may be used.
- Initialize the system and calibrate the device and measuring system (parallel plate, d = 25 mm). After measuring the zero gap, begin loading the PDMS sample.
- As soon as the PDMS elastomer and crosslinking agent have been mixed, pipet the mixture onto the bottom plate of rheometer.
- Move the spindle down to completely contact the top of the PDMS sample.
- Carefully trim the loaded sample excess from the bottom plate.
- In a strain sweep test, apply increasing strains for each composition with different crosslinking density to ensure the polymer structure remains in the linear viscoelastic regime during all shear measurements.
NOTE: Strain values relevant for cell studies are typically in the range of 0.1-10%. We have found PDMS to be linear up to approximately 100% strain. - Measure the dynamic shear storage modulus (G'), and loss modulus (G′′) of the PDMS network in a time sweep test with frequency of 1 Hz and oscillatory shear strain of 0.5% at 100 °C.
- To determine the viscoelasticity and time dependency of the final PDMS network, apply a frequency sweep test with frequency ranging 0.1-100 Hz and oscillatory shear strain of 0.5%.
Representative Results
Before addition of TGF-β, a confluent monolayer of cells has a cobblestone like shape and is tightly packed. Upon TGF-β treatment, cells become more elongated in morphology, enlarging the cell area and acquiring a more mesenchymal phenotype. Utilizing the multi-well device fabricated with soft PDMS elastomers, the physical properties of cells in a total 17 different conditions were studied. The cells were treated with four different TGF-β concentrations (0.5, 1, 2, and 4 ng/mL) and four different incubation times (12, 24, 48, 96 h), and these results are summarized in Figure 5. The cells treated with TGF-β applied larger traction stresses and strain energies than the cells cultured without TGF-β. Cells incubated with TGF-β for 96 h showed the largest traction stresses and strain energies. The cells applied larger stresses and strain energies when treated with higher concentration of TGF-β. The difference in tractions and strain energies were more distinct at longer incubation times.
The surface of the substrate needs to be smooth and uniformly coated with ligands, such as fibronectin or collagen. Scratched surface and/or non-uniform coating of ligands may lead to improper cell attachment, resulting in inaccurate traction measurement. Figure 6 shows the localized traction stresses due to the non-uniform substrate surface.
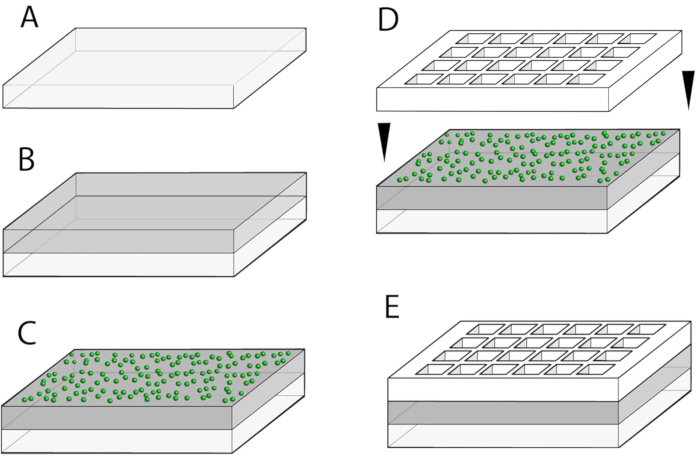
Figure 1: Overview of multi-well plate fabrication. (A) A custom-cut glass slide is the starting point. (B) The glass slide is coated with a thick (~100 µm layer of PDMS). (C) A layer of fiduciary beads (shown in green) are then spin-coated in a ~1 µm thick layer on top of the previous layer. (D) The multi-well divider is carefully placed on top of the fiduciary bead layer. (E) The complete multi-well plate is assembled and ready for use or storage. Please click here to view a larger version of this figure.
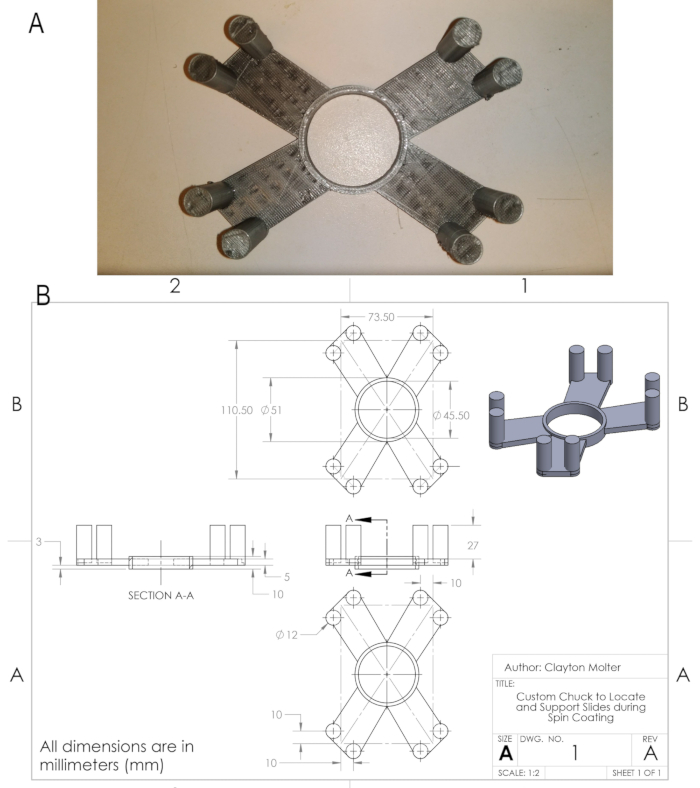
Figure 2: Custom chuck for spin-coater. To prevent the bottom glass (where PDMS is coated) from flying off of the standard spin-coater chuck, a custom-made holder is placed onto the chuck. (A) A custom-made holder for the spin-coater chuck. (B) Engineering drawing of the holder with all the dimensions. Please click here to view a larger version of this figure.
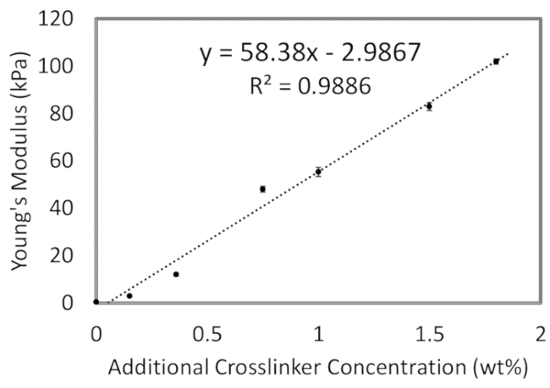
Figure 3: Substrate stiffness with changing crosslinker concentration (wt%). PDMS mixtures are prepared as described, with the additional crosslinker percentage increasing the elastic modulus until a max of 100 kPa at 1.8 weight percent (wt%) crosslinker. As a guide, the modulus as a function of crosslinker is fit linearly, which can be used by researchers to formulate their own mixture at a particular desired modulus within this range. Please click here to view a larger version of this figure.
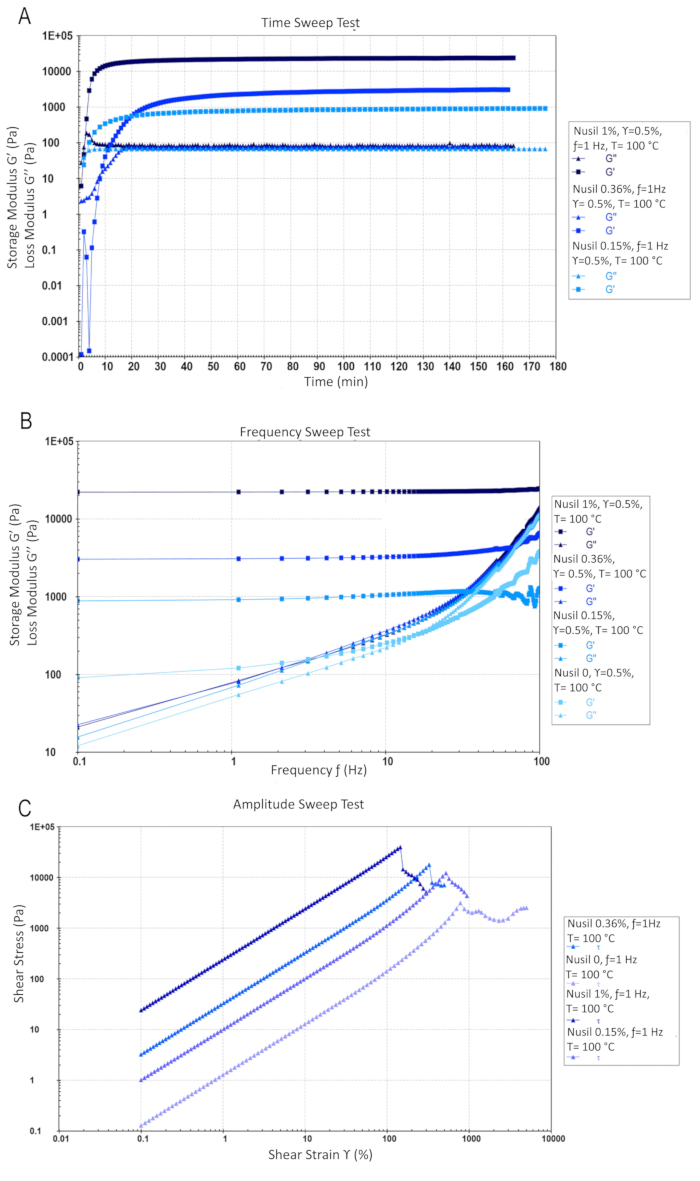
Figure 4: Rheology of PDMS elastomer containing 0, 0.15, 0.36 and 1 wt% of crosslink agents at a temperature of 100 °C. Triangle symbols indicate loss modulus (G'') and square symbols indicate storage modulus (G'). (A) Oscillatory time sweep at frequency of 1 Hz and shear strain of 0.5% during gelation. (B) Oscillatory frequency sweep of PDMS network at shear strain of 0.5%. (C) Strain sweep of PDMS network at frequency of 1 Hz. All data points were acquired in triplicate. Please click here to view a larger version of this figure.
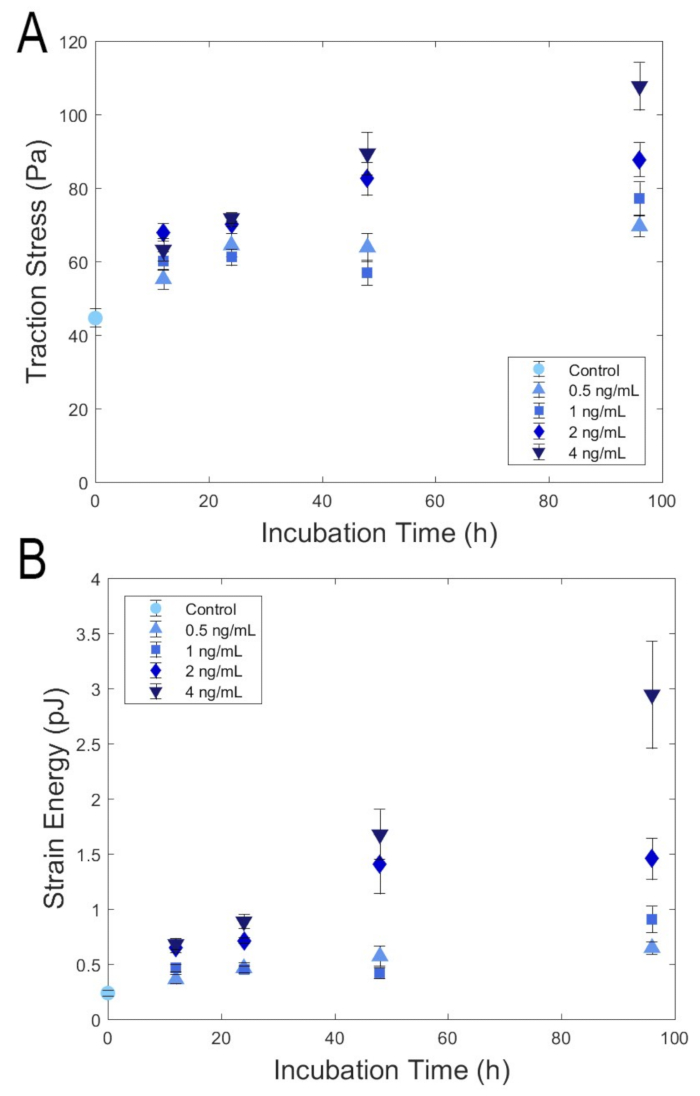
Figure 5: Representative results utilizing traction stress device with multi-well format. Monolayer of cells in different conditions were cultured in a multi-well device to measure contractility and strain energy. (A) Graph of traction stresses with increasing TGF-β incubation time for different TGF-β concentrations.Traction stresses increased with increasing TGF-β concentrations and incubation time. All data are statistically significant with respect to control (i.e., no TGF-β) except the following: 12 h – 0.5 ng, 12 h – 1 ng, 48 h – 1 ng. (B) Graph of strain energy with increasing TGF-β incubation time for different TGF-β concentrations. Strain energies increased with increasing TGF-β concentrations and incubation time. Sample sizes range from n = 7 to n = 15. All data are statistically significant with respect to control (i.e., no TGF- β) except the following: 12 h – 0.5 ng, 12 h – 1 ng, 24 h – 0.5 ng, 24 h – 1 ng, 48 h – 1 ng. Statistical significance was determined using the Kruskal Wallis test, which does not assume a normal distribution. Please click here to view a larger version of this figure.
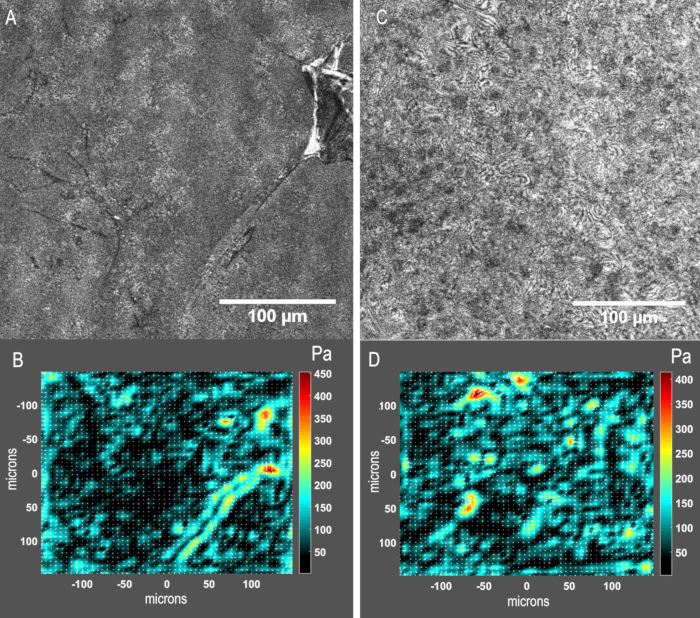
Figure 6: Representative results from a sub-optimal (A, B) and a satisfactory (C, D) experiment. (A) Reflection image of the substrate surface. Undesired contaminants, which can include dust or fibers, or material defects such as scratches are present on the substrate surface.(B)Stress map of cell contractility: due to the non-uniform surface, irregular and inaccurate traction stresses are observed around the objects.(C)Reflection image of the substrate surface. No apparent contaminants are visible.(D)Stress map of cell contractility: no artifact discontinuities are visible. Please click here to view a larger version of this figure.
| Crosslinker concentration (wt%) | G' (Pa) | Standard Deviation | E (kPa) | Standard Deviation |
| 0 | 0.135 | 0.014 | 0.405 | 0.043 |
| 0.15 | 1 | 0.1 | 3 | 0.35 |
| 0.36 | 4.027 | 0.245 | 12.081 | 0.73 |
| 0.75 | 16.01 | 0.49 | 48.03 | 1.2 |
| 1 | 18.44 | 0.989 | 55.32 | 1.94 |
| 1.5 | 27.638 | 0.93 | 82.91 | 1.64 |
| 1.8 | 33.986 | 0.88 | 101.94 | 1.088 |
| 2 | 33.36 | 0.67 | 100.08 | 1.1 |
Table 1:Substrate stiffness with changing crosslinker concentration (wt%). By changing additional crosslinker concentrations, PDMS substrates of the desired Young's moduli are fabricated. PDMS shear moduli were measured with a rheometer and Young's moduli were determined. For each data point, 3 independent preparations were done and the standard deviation is given.
Discussion
For the success of this method, it is critical to have a uniformly coated sample with a constant thickness of approximately 100 µm. The modulus should be carefully chosen to examine the physical significance of the biological system of interest. When fabricating a top layer, the concentration of the fiducial fluorescent particles should be optimized for accurate analysis of displacement and traction stress. Analyzing isolated single cells requires a denser fiduciary layer than measuring confluent monolayers. Additionally, the surface of the substrate should have stable and uniform coating of adhesion molecules such as collagen, fibronectin, and laminin to ensure proper attachment of the cells to the substrate surface. Particular care must be taken when attaching the multi-well divider; it should be placed without sliding to prevent sealing PDMS glue from being smeared on the culture surface, and the outer edge must be carefully aligned with the glass dimensions to prevent any leaks on the border wells.
To prevent wells from being leaky, an adequate amount of PDMS glue needs to be applied to the well-divider to hold it in place. However, use of excessive amounts will cover cell culture surface.
Sufficient volume of PDMS needs to be added during the spin-coating procedure. When curing, the oven and racks need to be level. Inadequate PDMS or an unleveled oven may lead to uneven PDMS thickness.
Bead density needs to be optimized for proper analysis. When the bead density is not adequate, concentration of the beads in fiduciary PDMS mixture needs to be increased. Additionally, an adequate amount of the mixture needs to be added for spin coating. To ensure proper fluorescent signals of the fiduciary bead particles, curing condition should be carefully monitored. Longer times and higher temperatures than the ones specified in this protocol may degrade the fluorescence. Improper protein coating on the surface of the sample may lead to poor cell adhesion. To prevent this, the surface needs to be cleaned and the expiration of reagents such as Sulfo-SANPAH and ligands needs to be checked. UV exposure time and strength should be optimized. Immunofluorescence or other fluorescent protein construct may be tested to ensure uniform ligand coating.
The device fabricated with this method and set of PDMS formulations is only applicable to substrates with Young's moduli ranging from 0.4 kPa to 100 kPa. Other moduli may be possible using alternative formulations, however, the current range spans a large set of physiologically relevant values. While this method is specifically for multi-well fabrication, it may also be used to prepare individual slides or other slide geometries, and in these instances further user troubleshooting may be necessary. In this embodiment, a relatively thick (1 mm) glass slide is employed, precluding common high numerical aperture (NA) objectives on an inverted microscope. While it is technically feasible to construct these multi-well TFM dishes using thinner glass, the fragility of these in processing and assembly may result in a high rate of breakage, and can run contrary to the higher-throughput approach. Furthermore, surface coating may be investigated further for wider applications of the device.
Utilizing a multi-well format, high-throughput experiments are possible. Multi-well format enabled us to examine the physical changes of NMuMG cells during EMT with TGF-β treatment with combinations of different concentrations and different incubation times in a single dish. Fabrication of traction stress devices with hydrogels such as polyacrylamide gel have several limitations. With the dominant ingredient being water, hydrogels are sensitive to salt concentrations (osmolarity), temperature, humidity, and pH changes. Changes in any of these properties can lead to the change in mechanical properties of the material, requiring extra care in using samples and interpreting results. Furthermore, having water, hydrogels are more susceptible to infections, requiring extra care. In contrast to water-based hydrogels, silicone-based PDMS is stable and inert. Once fabricated, it can be stored at room temperature indefinitely without requiring any special handling or storage conditions.
The impermeable nature of PDMS allows us to fabricate a monolithic substrate, ensuring that all wells are truly identical in substrate thickness and composition, while allowing unique biochemistry to be applied in the media, or different cells to be utilized in each well. A hydrogel-based surface allows diffusive factors to permeate and travel through it, necessitating adding the hydrogel to wells that are otherwise partitioned to prevent cross-contamination.
This method allows researchers to study cell contractility, a basic and ubiquitous aspect of cell biology, in a high-throughput manner, enabling more efficient and reproducible data acquisition. This helps to translate Traction Force Microscopy into Contractile Force Screening, allowing researchers to truly use contractility as a metric for cell activity, or efficacy of a compound. As a methodology, this has broad applications across biophysical and biomedical sciences, from understanding the physics of cells, to characterizing and testing pharmaceutical compounds against standardized cell types, or perhaps highly specialized individual cells for personalized medicine.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank Tom Kodger, Michael Landry, and Christopher J. Barrett for assistance with bead synthesis. A.J.E. acknowledges Natural Sciences and Engineering Research Council grants RGPIN/05843-2014 and EQPEQ/472339-2015, Canadian Institutes of Health Research grant no. 143327, Canadian Cancer Society grant no. 703930, and Canadian Foundation for Innovation Project #32749. R. Krishnan acknowledges National Institutes of Health grant no. R21HL123522 and R01HL136209. H.Y. was supported by Fonds de recherche Santé Québec, and Fonds de recherche Nature et Technologies Québec. The authors thank Johanan Idicula for assistance with the video and manuscript and Zixin He for assistance in preparing the video.
Materials
| Plate | |||
| GEL-8100 | Nusil Technology | GEL-8100 | High Purity Dielectric, Soft Silicone Gel kit |
| Dow Corning Sylgard 184 Silicone Encapsulant Clear 0.5 kg Kit | Ellsworth Adhesives | 184 SIL ELAST KIT 0.5KG | curing agent |
| Custom Cut Glass | Hausser Scientific Company | 109.6mm± x 72.8mm± x 1mm thickness | |
| Target 2TM Nylon Syringe Filter | ThermoFisher Scientific | F2513-4 | |
| 96-well Stripwell Egg Crate Strip Holder | Corning | 2572 | |
| Polystyrene Universal Microplate Lid With Corner Notch | Corning | 3099 | |
| Ethyl alcohol | Greenfield Global | P016EA95 | 0.95 |
| 2-Propanol | Sigma-Aldrich | 190764 | ACS reagent, ≥99.5% |
| Surface Coating | |||
| Sulfo-SANPAH Crosslinker | Proteochem | c1111-100mg | |
| Fibronectin bovine plasma | Sigma-Aldrich | F1141-1MG | solution, sterile-filtered, BioReagent, suitable for cell culture |
| PBS, 1X | Wisent | 319-005-CL | pH 7.4, without calcium and magnesium |
| DMSO | Sigma-Aldrich | 472301 | |
| Cell Culture | |||
| DMEM, 1X | Wisent | 319-005-CL | 4.5g/L glucose, with L-glutamine, sodium pyruvate and phenol red |
| FBS (Fetal Bovine Serum) | Wisent | 080-150 | Premium Quality, Endotoxin <1, Hemoglobin <25 |
| HEPES | Wisent | 330-050-EL | 1M, free acid |
| Human Insulin Recombinant | Wisent | 511-016-CM | USP grade |
| Penicillin-Streptomycin Solution | Wisent | 450-201-EL | 100 X, sterile filtered for cell culture |
| L-Glutamine solution | Wisent | 609-065-EL | 200mM solution, sterile filtered for cell culture |
| Amphotericine B | Wisent | 450-105-QL | 250μg/ml, sterile filtered for cell culture |
| Recombinant Human TGF-β1 | Peprotech | 100-21 | HEK293 Derived |
| Acetic acid | Sigma-Aldrich | 537020 | Glacial, ≥99.85% |
| Cictric acid | Sigma-Aldrich | 251275 | ACS reagent, ≥99.5% |
| NMuMG | ATCC | CRL-1636 | Mouse Mammary Gland Cell Line |
| Sodium azide | Fisher Schientific | AC190385000 | 99%, extra pure, ACROS Organics |
| Potassium hydroxide | Sigma-Aldrich | 221473 | ACS reagent, ≥85%, pellets |
| TritonX-100 | Sigma-Aldrich | X100 | laboratory grade |
| Bead Synthesis | |||
| 1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) | Sigma-Aldrich | 468495-100MG | 97% |
| Methyl methacrylate | Sigma-Aldrich | M55909-500ML | contains ≤30 ppm MEHQ as inhibitor, 99% |
| Inhibitor Remover | Sigma-Aldrich | 306312-1EA | Prepacked column for removing hydroquinone and monomethyl ether hydroquinone |
| Methacryloxylpropyl Terminated Polydimethylsiloxane | Gelest | DMS-R31 (25,000g/mol) | Polydimethylsiloxane stabilizer, 25,000g/mol, 1,000 cSt |
| 2,2′-Azobis(2-methylpropionitrile) (AIBN) | Sigma-Aldrich | 441090-25G | 98% |
| Hexane | Sigma-Aldrich | 296090-2L | anhydrous, 95% |
| Hexane, mixture of isomers | Sigma-Aldrich | 227064-1L | anhydrous, ≥99% |
| Whatman qualitative filter paper, Grade 1 | Sigma-Aldrich | WHA1001055 | circles, diam. 55 mm, |
| Equipment | |||
| Laurell WS-650Mz-23NPPB | Laurell Technologies | ||
| UVP Handheld UV Lamp Model UVGL-58 | VWR | 21474-622 | |
| Rheometer | Anton Paar | MCR 302 WESP | |
References
- Harris, A., Wild, P., Stopak, D. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science. 208 (4440), 177-179 (1980).
- Oliver, T., Dembo, M., Jacobson, K. Traction forces in locomoting cells. Cell Motil Cytoskeleton. 31 (3), 225-240 (1995).
- Dembo, M., Wang, Y. L. Stresses at the Cell-to-Substrate Interface during Locomotion of Fibroblasts. Biophysical Journal. 76 (4), 2307-2316 (1999).
- Yoshie, H., Koushki, N., et al. Traction Force Screening Enabled by Compliant PDMS Elastomers. Biophysical Journal. 114 (9), 2194-2199 (2018).
- Park, C. Y., Zhou, E. H., et al. High-throughput screening for modulators of cellular contractile force. Integrative biology quantitative biosciences from nano to macro. 7 (10), 1318-1324 (2015).
- Kraning-Rush, C. M., Califano, J. P., Reinhart-King, C. A. Cellular traction stresses increase with increasing metastatic potential. PLoS ONE. 7 (2), e32572 (2012).
- Agus, D. B., et al. A physical sciences network characterization of non-tumorigenic and metastatic cells. Scientific Reports. 3 (1), 1449 (2013).
- Guo, M., Ehrlicher, A. J., et al. Probing the Stochastic, Motor-Driven Properties of the Cytoplasm Using Force Spectrum Microscopy. Cell. 158 (4), 822-832 (2014).
- Ngan, E., Northey, J. J., Brown, C. M., Ursini-Siegel, J., Siegel, P. M. A complex containing LPP and alpha-actinin mediates TGFbeta-induced migration and invasion of ErbB2-expressing breast cancer cells. Journal of Cell Science. 126 (Pt 9), 1981-1991 (2013).
- Klein, S. M., Manoharan, V. N., Pine, D. J., Lange, F. F. Preparation of monodisperse PMMA microspheres in nonpolar solvents by dispersion polymerization with a macromonomeric stabilizer. Colloid & Polymer Science. 282 (1), 7-13 (2003).

