Multicolor 3D Printing of Complex Intracranial Tumors in Neurosurgery
Summary
The protocol describes the fabrication of fully colored three-dimensional prints of patient-specific, anatomical skull models to be used for surgical simulation. The crucial steps of combining different imaging modalities, image segmentation, three-dimensional model extraction, and production of the prints are explained.
Abstract
Three-dimensional (3D) printing technologies offer the possibility of visualizing patient-specific pathologies in a physical model of correct dimensions. The model can be used for planning and simulating critical steps of a surgical approach. Therefore, it is important that anatomical structures such as blood vessels inside a tumor can be printed to be colored not only on their surface, but throughout their whole volume. During simulation this allows for the removal of certain parts (e.g., with a high-speed drill) and revealing internally located structures of a different color. Thus, diagnostic information from various imaging modalities (e.g., CT, MRI) can be combined in a single compact and tangible object.
However, preparation and printing of such a fully colored anatomical model remains a difficult task. Therefore, a step-by-step guide is provided, demonstrating the fusion of different cross-sectional imaging data sets, segmentation of anatomical structures, and creation of a virtual model. In a second step the virtual model is printed with volumetrically colored anatomical structures using a plaster-based color 3D binder jetting technique. This method allows highly accurate reproduction of patient-specific anatomy as shown in a series of 3D-printed petrous apex chondrosarcomas. Furthermore, the models created can be cut and drilled, revealing internal structures that allow for simulation of surgical procedures.
Introduction
Surgical treatment of skull base tumors is a challenging task requiring precise preoperative planning1. Multimodal imaging using computed tomography (CT) and magnetic resonance imaging (MRI) provides the surgeon with information about the patient's individual anatomy. In clinical practice, this diagnostic information is visualized by displaying a series of two-dimensional (2D) cross-sections representing different aspects of the anatomy (e.g., CT for visualization of bone, CT angiography for vessels, MRI for soft tissue).
However, especially for beginners, medical students, and patients, understanding the complex relations of the different 3D structures to the cross-sectional images is challenging. Beside cadaveric studies2, this problem could be addressed by establishing real-size anatomical models of individual pathologies, displaying anatomical structures in different colors3.
Thanks to technical advances in the last years, 3D printing technologies allow cost-effective construction of complex shapes4,5. Therefore, this technique offers the possibility to construct patient-specific anatomical models that are tangible, clearly depict spatial relations, and can be used for surgical planning and simulation. Especially in rare and complex cases such as petrous apex chondrosarcomas, preoperative simulation of tumor removal in an individual case might help to improve the self-confidence of the surgeon and the patient outcome.
Common FDM-printing (filament deposition modelling) techniques only allow for creating objects with a closed surface in one or a limited variety of colors6. To provide a model for surgical simulation that contains various complex shaped anatomical structures mainly nested inside each other, fully volumetrically colored 3D prints are needed. This allows for a successive removal of tissue layers until an internal structure is revealed.
Plaster-based color 3D binder jetting is a technique able to produce the required multicolor models7. Whereas in its standard configurations only the surface of an object can be colored, herein a modified technique is described to ensure volumetric application of color to internal anatomical structures.
To demonstrate this technique, cases of patients with skull base chondrosarcomas were selected as an example. Chondrosarcomas account for 20% of all neoplasia in the skeletal system, mostly located in the long bones. Primary skull base chondrosarcomas are a rare condition responsible for 0.1–0.2% of all intracranial tumors8. Mainly located at the petrous apex, these tumors grow in a complex anatomical environment involving pivotal structures such as the internal carotid artery, the optic and other cranial nerves, as well as the pituitary gland. Treatment of these neoplasms is mainly focused on a total surgical resection, because adjuvant therapies alone (e.g., radiation) are not effective enough9.
Due to the complexity and rarity of this tumor entity, preoperative surgical simulation in a 3D printed skull model might help to better visualize and understand the anatomy and to assist the surgeon achieve complete resection. As shown by others10,11 3D printing of patient-specific models improves both residents' and experienced neurosurgeons' understanding of complex neuroanatomy.
However, creating such individualized models from medical imaging data requires skills in image segmentation, 3D modelling, and 3D printing, especially when anatomical structures are to be printed in different colors. This manuscript intends to make fabrication of the described anatomical models more accessible for others by providing a detailed protocol for converting medical imaging data into virtual 3D models and for the fabrication of multicolored 3D objects.
The workflow mainly consists of four parts: 1) segmentation of medical imaging data and creation of a virtual 3D model; 2) preparation of the virtual 3D model for multicolor 3D printing; 3) preparation for volumetric coloring of selected parts; and 4) 3D printing and post processing.
Protocol
The protocol was approved by the responsible local ethics committee (Ethikkommission der Landesärztekammer Rheinland-Pfalz, Deutschhausplatz 3, 55116 Mainz, Germany). All institutional guidelines for the care and use of patient data were followed.
1. Segmentation of medical imaging data and creation of a virtual 3D model
NOTE: The software we used for segmentation was Amira 5.4.5. The segmentation process can also be accomplished by using open source software (e.g., 3D Slicer, https://www.slicer.org/)
- Use imaging data with high spatial resolution (e.g., a slice thickness of 1 mm or less). Here, a cranial CT dataset with a slice thickness of 0.5 mm and additional MRI data with a slice thickness of 1 mm were used. Use CT data for the segmentation of bone, contrast enhanced T1 MRI images for the segmentation of tumor and neural structures, as well as time-of-flight (TOF) images for vessels.
- Download the DICOM files on the computer and open the segmentation software. Import the files of the different imaging modalities and select the folder with the imaging data.
- Click on the CT images and connect them with a volume rendering module (Volren). Choose Specular for a more realistic rendering and adjust the color transfer slider to visualize bone only. Continue by importing the MRI sequences and connect them to a volume rendering module as well.
- Registration
- Because the MRI and CT images do not overlap, it is necessary to fuse the different imaging data. Therefore, right click on the MRI dataset and choose Compute > Affine Registration. Choose Reference by clicking on the white square of the module, then drag the cursor onto the CT.
- In the Registration Module's properties leave all settings on default and click on Align Centers, followed by clicking 등록. The two different imaging datasets are now fused. Repeat this step for all further imaging datasets.
- Verification of matching accuracy:
- Check the matching accuracy by hiding the volume renderings (click on the orange square of the module) and adding an OrthoSlice module to the MR images. Click on the white triangle and choose Colorwash. Next click on the white square, choose Data and connect this port with the CT data by dragging the mouse onto it.
- Adjust the color slider to visualize the neural structures superimposed to the bony skull structures. Check for any misalignments by toggling the weight factor slider while looking at the border between the skull and brain surfaces as well as the ventricles. Repeat this procedure on different slices in coronal and sagittal directions.
- Volumetric editing
- Deactivate the OrthoSlice module's visibility and reactivate volume rendering of the CT. Go to the CT data and look for the lowest value in the dataset, in this case -2,048.
- Next add a Volume Edit module, connect the Volren module with the output data, and set the Padding Value to -2,048.
- Click on Cut Inside and mark the region to be removed in the 3D viewport.
NOTE: It is important to avoid overlapping with parts not intended to be removed. - In this example, parts of the mandible bone and the upper cervical vertebrae were removed.
- Segmentation of bone
- Next, the remaining bone must be segmented and converted into a surface mesh. To do so, click on the Segmentation Editor, choose the modified CT image sequence, and add a new Labelset by clicking on 신규.
- Now choose Threshold as a segmentation option. Set the lower slider to a value of ~250 in case of a CT. Ensure that thin bone structures such as the temporal bone or the upper orbital region are selected in the preview. Otherwise, adjust the lower threshold but avoid selecting any soft tissue.
- Next click on Select and finally add the selection to the labelset (by clicking on the red plus icon).
- Return to the Pool View. A new labelset has been created for the CT. Right click and choose Compute > Surface Gen, check the Compactify option, and click Apply.
- Finally, add a SurfaceView module and adjust the color of the generated mesh.
- Segmentation of other structures
- Add other relevant structures by repeating the previous steps. In case of the tumor, manual segmentation was used rather than a thresholding operation.
- To perform manual segmentation, go to the Segmentation Editor, and choose the manual segmentation option (brush icon) to mark structures such as the tumor in every slice. Finally add the selection again by clicking onto the plus icon. Thus, the tumor, the optic nerve, and the intracranial vessels will be segmented and added to the model.
- Exporting meshes
- Finally export the generated meshes in the STL format by right clicking on the mesh and clicking on Save. Choose binary STL as the file format.
2. Preparation of the virtual 3D model for multicolor printing
NOTE: The software used for print preparation in this protocol is Netfabb Premium 2019.0. Autodesk offers free use of this software in its educational program.
- Import data and perform automatic repair.
- Open the print preparation program and import the meshes generated in the previous steps as new parts. Check Automatic Repair and click Import.
- Deleting small loose parts
- Select the skull and split its shells into parts by clicking on Modify > Split Shells into Parts. This separates any loose objects not connected to the skull bone.
- Select the skull bone and toggle its visibility off.
- Now select all other parts and delete them.
- Toggle the skull's visibility on again.
- Repeat this step for all other objects.
- Remove overlapping regions.
NOTE: In some regions, such as the tumor inside the petrous apex of the skull, the geometries of both objects intersect each other. To avoid printing errors, it is necessary to remove such intersections.- Select the two intersecting objects and click on Boolean Operations.
- Move the object to be subtracted from the other one to the red side of the list and click on Apply. Now the two objects are clearly separated. This should be checked by toggling their visibility.
- Repeat these steps to allow the tumor as well as the artery inside the tumor to become clearly separated from each other.
- Add supporting structures where needed.
- In case of the basilar artery, additional supports are needed to prevent the object from being a loose part after printing.
- Add a new object, in this case a cylinder (File > Part Library), and adjust its dimensions and subdivisions as needed.
- Place the cylinder to completely intersect with the skull and the vessel geometry.
- Now perform the Boolean operation again to subtract the parts within the bone and the blood vessel.
- Repeat this step to add more supports where needed (e.g., the optic nerve).
3. Preparation for volumetric coloring of selected parts
NOTE: To allow volumetric coloring of certain parts it is necessary to generate not only one surface shell but many subshells (additional surfaces) inside the object.
- Select the tumor, in this case, and generate a new shell from it (right click > Modify > Generate Shell).
- Set a shell thickness of 0.3 mm in the Inner Offset Mode with an accuracy of 0.15 mm and Apply. Select the Keep Original Part checkbox. This generates an inner shell with a distance of 0.3 mm to the original surface.
- Select the outside surface of both shells and generate a new shell from it. Select a shell thickness of 0.25 mm in Hollow Mode with an accuracy of 0.15 mm. Also select the Remove Original Part checkbox. This generates a space of 0.05 mm between the two adjacent shells.
- Repeat steps 3.1–3.3 so that multiple inner shells with constant thicknesses and invariant offsets are created.
NOTE: It is recommended to use a shell thickness of 0.35–0.25 mm as well as an offset of 0.1–0.05 mm to achieve smooth volumetric coloring. - Repeat steps 3.1–3.4 with all other objects, such as the blood vessels.
4. Coloring and exporting of the 3D model
NOTE: The coloring of all parts of the model, including the distinct nested shells, is done using the Netfabb software.
- Select a part to be colored in the Parts Menu on the left side. Double click the Texture and Color Mesh icon. Choose a color by clicking the color bar on the right side. In the top menu left click on the Paint on Shells icon. Subsequently left click on the model displayed in the screen center. Finally left click on the Apply Changes box in the lower right corner. Be sure to confirm the Remove Old Part selection.
- Repeat these steps with all other objects and shells, respectively.
- Export all objects. Select all objects to be printed, including supports and inner shells, and export them as individual files. Be sure to choose the VRML (WRL) format, because the STL format is not able to transport the color information.
5. Printing and post processing of the 3D model
- Set up the 3D printer
NOTE: The 3DPrint Software (Version 1.03) was used to control the ZPrinter 450 binder jetting machine.- Open the software and import the colored VRML files by clicking on Open and choosing all relevant data. Click on the Open button in the lower right window corner. In the subsequent window choose millimeters as units. Make sure to check the Keep Position and Orientation as well as the Apply Settings to All Files boxes. Finally choose Z151 as the material type. Click on the Next button.
- For positioning the 3D objects inside of the build volume, mark all objects by pressing the <STRG>+A key.
- In the upper left window, representing the XY view of the build volume, click and drag the marked objects to the center. In the lower left window, representing the XZ view of the build volume, click and drag the objects in the middle of the lower part above the yellow line.
- If a whole skull model is printed, be sure that the opening is facing upwards. If isolated small models are printed, be sure to align delicate parts such as vessels with the XY plane, because this orientation will increase the strength of the respective parts.
- Check the correct orientation of the models by clicking and moving them in the window on the right-hand side.
- To prepare for the build process, click on the Setup icon in the upper menu. Make sure that the correct Material Type is selected and that the Layer Thickness is set to 0.1 mm. The Bleed Compensation should be checked and the Print in Monochrome option unchecked.
- To start the printing process, click on the Build icon in the upper menu. In the subsequent window choose Entire Build and click the OK button. Be sure that in the following Printer Status dialogue all listed items are set correctly and that the printer is online. Then, click on the Print button in the lower part of the dialogue box.
- Post processing of the model
NOTE: Always wear a laboratory coat, gloves, eye protection, and a mask while handling the loose powder and the hardening solution. Always work in a well-ventilated area.- Unpacking
- After the print is finished, unpack the model by carefully removing the loose powder with the integrated vacuum cleaner. It is important not to directly contact the model with the suction tube to prevent thin structures from breaking apart.
- Remove the model and clean it by applying pressurized air as well as cleaning it with a soft brush. Thicker, more stable, parts of the model can additionally be ground with a harder brush. This optional step allows for a smoother surface finish. Keep in mind that in this state the model is still very fragile.
- Infiltration
- Put the model inside a plastic tub.
- Infiltrate it carefully with the hardening solution until no whitish areas are visible.
- The surplus solution has to be removed with pressurized air and disposable paper towels to maintain all the surface details.
- Let the model cure for several hours until it is completely dry.
- Unpacking
Representative Results
Eight patients with chondrosarcoma of the petrous apex were selected for the study and virtual 3D models were created, each containing bone, tumor, vessels, pituitary gland, and optic nerve crossing. Three models underwent multicolored 3D printing using the plaster-based color 3D binder jetting technique (Figure 1A1,A2). Additionally, a single tumor with an internal artery was created (Figure 1B1) to show the benefits of volumetric coloring compared to surface coloring (Figure 1B2,B3).
These models were used to demonstrate the simulation of a surgical approach (e.g., burr hole creation) and tumor resection. This printing technique allowed for combining anatomical structures derived from different imaging modalities into one single object.
The plaster material had bone-like properties and could be easily drilled without melting. Thus, it was feasible to use it to simulate a surgical access route. After the hardening procedure it was stable enough to reproduce even fragile structures such as the intracerebral vessel tree.
The ability to color the entire volume of the model allowed an object's internal structure, such as the internal carotid artery travelling through the tumor, to be clearly visualized. By removing layers of tumor with the drill, the red artery was gradually revealed during the surgical simulation.
To prove the accuracy of the technique, 3D models were scanned in a computer tomograph. The models created for printing were superimposed to these scans. A deviation mapping was created, and the accuracy was determined in 50 randomly chosen surface points. A mean deviation of 0.021 mm demonstrates the high accordance of the 3D print compared to the original data.
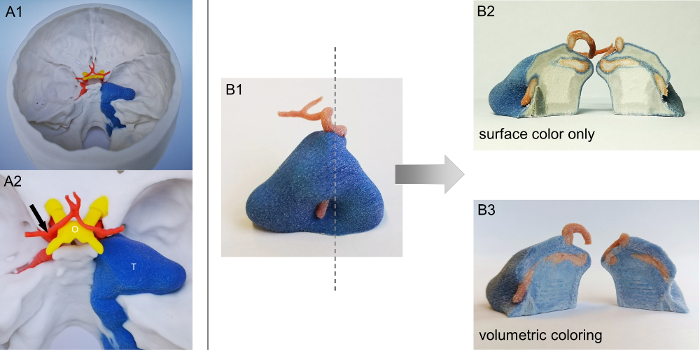
Figure 1: Volumetric vs. surface colored 3D prints. A1. Exemplary full color 3D print of a patient with chondrosarcoma at the right petrous apex. A2. Detailed view of anatomical structures (arrow = internal carotid artery bifurcation; O = optic nerve chiasm; T = tumor). B1. Blood vessel crossing the tumor volume and cross-sectional level (dotted line). B2. The conventional multicolor printing technique reveals color only at the surface. B3. The modified technique produces volumetrically colored objects suitable for advanced surgical simulation. Please click here to view a larger version of this figure.
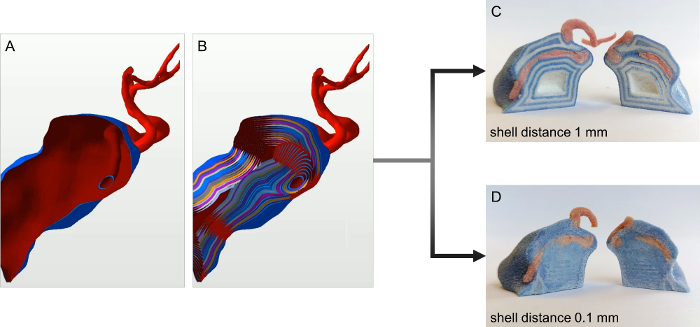
Figure 2: Workflow for volumetric coloring of 3D prints. A. Virtual 3D model of a tumor with a blood vessel crossing its volume without inner shells. B. A tumor and a blood vessel with multiple inner shells (distance 0.05 mm). C. An example of a high shell distance (1 mm). The single layers of colored and white shells are still visible. D. An example of small shell distance (0.1 mm). The object's inner volume is completely colored. Please click here to view a larger version of this figure.
Discussion
The therapy of intracranial chondrosarcoma is mainly based on complete surgical removal. Often located on the petrous apex, this tumor is close to important structures such as the internal carotid artery, the optic nerve, and the pituitary gland. Therefore, planning the surgical trajectories is a crucial step prior to surgery. Multicolor 3D printing allows for fusion of these structures, each derived from different imaging modalities, into a single object.
During the preparation for 3D printing it is important to carefully select adequate imaging data. High resolution images with a small slice thickness are well suited for 3D reconstruction and smooth transitions, whereas high slice thicknesses will produce coarse, uneven objects. Another critical step of the method is to avoid any intersections of two neighboring objects such as tumor and skull bone. Therefore, Boolean operations have to be performed to subtract one object from the other.
To allow for volumetric coloring it is necessary to create onion shell-like subsurfaces inside an object (Figure 2A,B). It is necessary to have a minimum distance between two adjacent surfaces of at least 0.1 mm to obtain smoothly colored objects (Figure 2D). If the distance chosen is above this value, the individual shells inside the object might become visible (Figure 2C). Attention should be paid to an increased color consumption of the 3D printer when using volumetric coloring. Furthermore, it is also important to check the model for any loose parts and add supports when necessary (e.g., the basilar artery).
The method can only produce stiff, plaster-like material that is not very durable. Especially without the hardening procedure, the model can be easily destroyed during the unpacking procedure. Thus, fragile elements such as blood vessels often tend to break apart.
The technique is also not suitable for simulation of soft tissue. To simulate brain tissue, for example, it might be necessary to either print it with a method that is able to produce soft and hard materials directly12,13 or to print molds that can be used to cast soft objects, such as silicone rubber14. In one test case, the latter method was used to simulate a soft tumor. The limitation of this last procedure was that although the silicone tumor was very flexible, it was necessary to have enough space to insert it into the 3D printed model. Furthermore, it was not possible to create inner structures, such as a blood vessel.
3D binder jetting is an additive manufacturing technique that assembles objects by partial hardening and coloring thin layers of plaster powder. Thus, it allows for printing a nearly unlimited range of colors, color transitions, and colored structures inside the volume of objects in one single process.
Compared to other printing techniques such as filament printers, which produce the lowest costs but only allow two or three colors at once, and Poly Jet printers that produce multicolor, multi- material objects but are very expensive, this technique offers a compromise at an affordable price. The mean material cost for a printed skull was about 150 €.
With this method it is possible to visualize even more abstract data such as filament fibers derived from MRI fiber tracking sequences or functional imaging depicting, for example, the brain speech area (e.g., Broca%s area).
Apart from surgical simulation, 3D printed, fully colored models of real patient anatomy can help improve the education of medical students or young physicians so they can better understand complex anatomical relations. It is also an important tool in patient education.
Disclosures
The authors have nothing to disclose.
Acknowledgements
Parts of this work have been presented as a poster at the annual meeting of the German Neurosurgical Society (DGNC) 2019 in Würzburg, Germany and as a short presentation at the annual meeting of the German Society for Computer and Robot Assisted Surgery (CURAC) 2019 in Reutlingen, Germany.
Materials
| 3D printer | 3D Systems (formerly Zcorp) | x | Zprinter Z450 |
| 3D printing software | 3D Systems (formerly Zcorp) | x | 3DPrint Software (Version 1.03) |
| Binder solution for cartridge | 4D Concepts GmbH, Groß-Gerau, Germany | 42-0100-7001 | VisiJet PXL Binder Cartridge clear 1 x ca. 1 Liter |
| Infiltration solution | 4D Concepts GmbH, Groß-Gerau, Germany | 42-0250-1090 | Color-Bond 90, 1 bottle, 454 g |
| Modeling Software for 3D print preparation | Autodesk, San Rafael, CA, USA | x | Netfabb Premium (Version 2019.0) |
| Print head for binder | 4D Concepts GmbH, Groß-Gerau, Germany | 42-0150-2010 | HP 11 print head (C4810A) |
| Print head for color | 4D Concepts GmbH, Groß-Gerau, Germany | 42-0150-2011 | HP 57 printhead C 6657 AE Tricolor |
| Printing powder | 4D Concepts GmbH, Groß-Gerau, Germany | 42-0050-2061 | VisiJet PXL Core Eco Drum ca. 14 kg – ca. 11,47 L |
| Segmentation software | Thermo Fisher Scientific, Waltham, MA, USA | x | Amira 5.4.5 |
References
- Frank, G., et al. The endoscopic transnasal transsphenoidal approach for the treatment of cranial base chordomas and chondrosarcomas. Neurosurgery. 59, 50-57 (2006).
- Wu, P., et al. Quantitative evaluation of different far lateral approaches to the cranio-vertebral junction using the microscope and the endoscope: a cadaveric study using a tumor model. Acta Neurochirurgica. 160, 695-705 (2018).
- Huang, X., et al. A small 3D-printing model of macroadenomas for endoscopic endonasal surgery. Pituitary. 22 (1), 46-53 (2018).
- Stone, J. J., Matsumoto, J. M., Morris, J. M., Spinner, R. J. Preoperative Planning Using 3-Dimensional Printing for Complex Paraspinal Schwannoma Resection: 2-Dimensional Operative Video. Operative Neurosurgery. 16 (3), 80 (2018).
- Scerrati, A., et al. A workflow to generate physical 3D models of cerebral aneurysms applying open source freeware for CAD modeling and 3D printing. Interdisciplinary Neurosurgery. 17, 1-6 (2019).
- Kamio, T., et al. Utilizing a low-cost desktop 3D printer to develop a “one-stop 3D printing lab” oral and maxillofacial surgery and dentistry fields. 3D Printing in Medicine. 4, 6 (2018).
- Kondo, K., et al. A neurosurgical simulation of skull base tumors using a 3D printed rapid prototyping model containing mesh structures. Acta Neurochirurgica. 158, 1213-1219 (2016).
- Awad, M., Gogos, A. J., Kaye, A. H. Skull base chondrosarcoma. Journal of clinical Neuroscience: Official Journal of the Neurosurgical Society of Australasia. 24, 1-5 (2016).
- Jones, P. S., et al. Outcomes and patterns of care in adult skull base chondrosarcomas from the SEER database. Journal of Clinical Neuroscience: Official Journal of the Neurosurgical Society of Australasia. 21, 1497-1502 (2014).
- Karakas, A. B., Govsa, F., Ozer, M. A., Eraslan, C. 3D Brain Imaging in Vascular Segmentation of Cerebral Venous Sinuses. Journal of Digital Imaging. 32 (2), 314-321 (2018).
- Dong, M., et al. Three-dimensional brain arteriovenous malformation models for clinical use and resident training. 의학. 97, 9516 (2018).
- Dolinski, N. D., et al. Solution Mask Liquid Lithography (SMaLL) for One-Step, Multimaterial 3D Printing. Advanced Materials. 30, 1800364 (2018).
- Coelho, G., et al. Multimaterial 3D printing preoperative planning for frontoethmoidal meningoencephalocele surgery. Child’s Nervous System: ChNS: Official Journal of the International Society for Pediatric Neurosurgery. 34, 749-756 (2018).
- Javan, R., Cho, A. L. An Assembled Prototype Multimaterial Three-Dimensional-Printed Model of the Neck for Computed Tomography- and Ultrasound-Guided Interventional Procedures. Journal of Computer Assisted Tomography. 41, 941-948 (2017).

