质谱导览基因组挖掘作为发现新天然产品的工具
Summary
这里建立了一个质谱仪指导基因组挖掘协议并对此进行描述。它基于基因组序列信息和LC-MS/MS分析,旨在促进从复杂的微生物和植物提取物的分子的识别。
Abstract
天然产品所覆盖的化学空间是巨大的,而且被广泛地未被识别。因此,需要方便的方法对其在性质和潜在的人类利益(例如,药物发现应用)的功能进行广泛的评估。该协议描述了基因组挖掘 (GM) 和分子网络 (MN) 的组合,这两种现代方法将全基因组测序中的基因簇编码注释与粗代谢提取物的化学结构特征相匹配。这是发现新的自然实体的第一步。这些概念,当一起应用,在这里被定义为MS引导基因组挖掘。在此方法中,主要组件以前被指定(使用 MN),并且与结构相关的新候选项与基因组序列注释(使用 GM)相关联。将GM和MN结合是一种有利可图的策略,旨在瞄准新的分子骨干或收获代谢特征,以便识别已知化合物中的类似物。
Introduction
二次代谢的研究通常包括筛选特定生物活动的粗提取物,然后对属于活性馏分的成分进行纯化、识别和表征。事实证明,这一过程是有效的,促进了若干化学实体的隔离。然而,现在这被认为是不可行的,主要是因为高的重新发现率。由于制药业在不了解特殊代谢物的作用和功能的情况下进行了革命,其鉴定是在实验室条件下进行的,没有准确代表自然1。今天,人们更好地了解了自然信号的影响、分泌,以及大多数目标在检测出的低浓度下的存在。此外,对这一过程的监管将有助于学术界和制药业利用这一知识。它还将有利于与沉默生物合成基因簇(BGCs)2相关的代谢物的直接分离研究。2
在此背景下,基因组测序的进步重新产生了对筛选微生物代谢物的兴趣。这是因为分析未发现的生物合成簇的基因组信息可以揭示基因编码在实验室条件下未观察到或生产的新化合物。许多微生物全基因组项目或草案今天可用,而且数量每年都在增长,这为通过基因组挖掘33、44发现新的生物活性分子提供了巨大的前景。
生物合成基因簇图集是目前最大的自动挖掘基因簇集合,作为联合基因组研究所(JGI IMG-ABC)2集成微生物基因组平台的组成部分。2最近,生物合成基因簇(MIBiG)标准化倡议的最低信息促进了BPC的手动再注释,提供了高度精心策划的参考数据集5。如今,大量的工具可用于计算挖掘遗传数据及其与已知次生代谢物的连接。还制定了不同的策略,以获取新的生物活性天然产品(即异构表达、靶基因删除、体外重组、基因组序列、同位素引导筛选[基因同位素方法]、操纵本地和全球监管机构、基于抗药性的采矿、培养独立采矿,以及最近MS引导/编码方法2,2、6、7、8、9。6,7,8,9,1010、11、12、13、14、15。,11,12,13,14,15
基因组挖掘作为一种单一策略,需要努力对单个或一小群分子进行批批;因此,在将新化合物列为隔离和结构阐明的优先位置的过程中仍然存在差距。原则上,这些方法每次实验只针对一种生物合成途径,从而导致缓慢发现速度。从这个意义上说,使用转基因和分子网络方法是天然产品研究的一个重要进步14,15。14,
液相色谱-质谱(LC-MS)的多功能性、准确性和高灵敏度使其成为复合鉴定的好方法。目前,几个平台已经投资算法和软件套件的无目标代谢组16,17,18,19,20。16,17,18,19,20这些程序的核心包括特征检测(峰值选取)21和峰值对齐,这允许在一批样本中匹配相同的特征并搜索模式。基于MS模式的算法22,23,23比较特征破碎模式,匹配MS2的相似性,生成共享结构特征的分子族。这些特征可以突出和聚类,赋予快速发现已知和未知的分子从复杂的生物提取物通过串联MS2,24,252,24,25的能力。因此,串联MS是一种多功能方法,用于同时获取大量数据中包含的几种化学型的结构信息。
全球天然产物社会分子网络(GNPS)26算法使用规范化片段离子强度来构造多维向量,其中相似性使用原成函数进行比较。不同父离子之间的关系在关系图表示形式中绘制,其中每个碎片都可视化为节点(圆),每个节点的相关性由边缘(线)定义。单一来源分子的全球可视化被定义为分子网络。结构上分裂分子,独特的碎片将形成自己的特定簇或星座,而相关分子聚集在一起。聚类化学型允许假设其生物合成起源类似的结构特征连接。
在建立BGC及其小分子产品之间的生物信息学联系时,将化学型到基因型和基因型到化学型的方法结合起来是强大的。因此,MS引导基因组挖掘是一种快速方法和低耗材料策略,它有助于在不同代谢和环境条件下,WGS揭示的一个或多个菌株的母离子和生物合成途径。
该协议的工作流程(图1)包括将WGS数据输入生物合成基因簇注释平台,如反SMASH28、29、30。28,29,30它有助于估计由基因组编码的化合物和化合物的种类。必须采用一种以生物合成基因簇为目标的策略,编码感兴趣的化学实体,并分析含有BGC的野生型菌株和/或异质菌株的培养提取物,以利用GNPS26、31,31的相似性生成聚类离子。因此,可以识别与目标 BGC 关联的新分子,并且数据库中不可用(主要是未知的类似物,有时以低脚点生成)。考虑用户可以为这些平台做出贡献,并且生物信息学和 MS/MS 数据的可用性正在迅速增加,从而推动有效计算工具和算法的不断发展和升级,从而指导复杂提取物与分子的有效连接, 这是相关的。
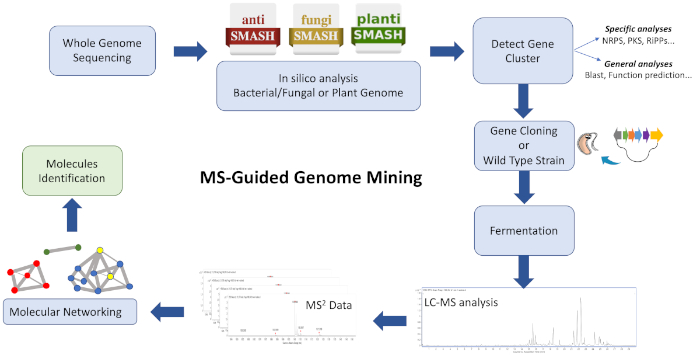
图 1:整个工作流的概述。如图所示,说明了在所述 MS 引导基因组挖掘方法中涉及的生物信息学、克隆和分子网络步骤,以识别新的代谢物。请点击此处查看此图形的较大版本。
该协议描述了一个快速而高效的工作流程,将基因组挖掘和分子网络结合为天然产品发现管道的起点。尽管许多应用能够可视化一个网络中MS可检测分子的组成和关联性,但此处采用了几种来可视化结构相似的聚类分子。利用这一策略,成功鉴定了在链球菌孢菌剂2042代谢提取物中观察到的新型环丙肽产物。在基因组挖掘的指导下,对华利霉素进行全生物合成基因簇编码被识别和克隆成生产者菌株链球菌素M1146。最后,在MS模式分子网络之后,MS检测到的分子与负责其生物成形的BPC相关。
Protocol
Representative Results
Discussion
该协议的最大优点是能够快速去复制代谢特征,并将基因组信息与MS数据桥接,以阐明新分子的结构,特别是结构模拟2。根据基因组信息,可以研究不同的天然产物化学型,如多基肽(PK)、非核糖类肽(NRP)和糖基化天然产物(GNP),以及神秘的BFC。 Metabolomic筛选可产生实验室条件下特定菌株产生的活性BGC轮廓和化学多样性的证据。因此,BGC 可以克隆,以直接生产与已知 BGC ?…
Disclosures
The authors have nothing to disclose.
Acknowledgements
这项研究的财政支助由圣保罗研究基金会-FAPESP(2019/10564-5, 2014/12727-5 和 2014/50249-8 到 L.G.O; 2013/12598-8 和 2015/01013-4 到 R.S.; 和 2019/08853-9 到 C.F.F.A. 。B.S.P.、C.F.F.A.和L.G.O.获得国家科学和技术发展委员会(CNPq)研究金(205729/2018-5、162191/2015-4和313492/2017-4)。L.G.O.还感谢《妇女参与科学》方案(2008年,巴西版)提供的赠款支持。所有作者都承认CAPES(高等教育人员改进协调)支持巴西的毕业后课程。
Materials
| Acetonitrile | Tedia | AA1120-048 | HPLC grade |
| Agar | Oxoid | LP0011 | NA |
| Apramycin | Sigma Aldrich | A2024 | NA |
| Carbenicillin | Sigma Aldrich | C9231 | NA |
| Centrifuge | Eppendorf | NA | 5804 |
| Chloramphenicol | Sigma Aldrich | C3175 | NA |
| Column C18 | Agilent Technologies | NA | ZORBAX RRHD Extend-C18, 80Å, 2.1 x 50 mm, 1.8 µm, 1200 bar pressure limit P/N 757700-902 |
| Kanamycin | Sigma Aldrich | K1377 | NA |
| Manitol P.A.- A.C.S. | Synth | NA | NA |
| Microcentrifuge | Eppendorf | NA | 5418 |
| Nalidixic acid | Sigma Aldrich | N4382 | NA |
| Phusion Flash High-Fidelity PCR Master Mix | ThermoFisher Scientific | F548S | NA |
| Q-TOF mass spectrometer | Agilent technologies | NA | 6550 iFunnel Q-TOF LC/MS |
| Sacarose P.A.- A.C.S. | Synth | NA | NA |
| Shaker/Incubator | Marconi | MA420 | NA |
| Sodium Chloride | Synth | NA | P. A. – ACS |
| Soy extract | NA | NA | NA |
| Sucrose | Synth | NA | P. A. – ACS |
| Thermal Cycles | Eppendorf | NA | Mastercycler Nexus Gradient |
| Thiostrepton | Sigma Aldrich | T8902 | NA |
| Tryptone | Oxoid | LP0042 | NA |
| Tryptone Soy Broth | Oxoid | CM0129 | NA |
| UPLC | Agilent Technologies | NA | 1290 Infinity LC System |
| Yeast extract | Oxoid | LP0021 | NA |
References
- Davies, J. Specialized microbial metabolites: functions and origins. The Journal of Antibiotics. 66 (7), 361-364 (2013).
- Ziemert, N., Alanjary, M., Weber, T. The evolution of genome mining in microbes – a review. Natural Product Reports. 33 (8), 988-1005 (2016).
- Zerikly, M., Challis, G. L. Strategies for the Discovery of New Natural Products by Genome Mining. ChemBioChem. 10 (4), 625-633 (2009).
- Gomez-Escribano, J. P., Bibb, M. J. Heterologous expression of natural product biosynthetic gene clusters in Streptomyces coelicolor: from genome mining to manipulation of biosynthetic pathways. Journal of Industrial Microbiology & Biotechnology. 41 (2), 425-431 (2014).
- Medema, M. H., et al. Minimum Information about a Biosynthetic Gene cluster. Nature Chemical Biology. 11 (9), 625-631 (2015).
- Lautru, S., Deeth, R. J., Bailey, L. M., Challis, G. L. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nature Chemical Biology. 1 (5), 265-269 (2005).
- Chiang, Y. -. M., et al. Molecular Genetic Mining of the Aspergillus Secondary Metabolome: Discovery of the Emericellamide Biosynthetic Pathway. Chemistry & Biology. 15 (6), 527-532 (2008).
- Huang, T., et al. Identification and Characterization of the Pyridomycin Biosynthetic Gene Cluster of Streptomyces pyridomyceticus NRRL B-2517. Journal of Biological Chemistry. 286 (23), 20648-20657 (2011).
- Udwary, D. W., et al. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proceedings of the National Academy of Sciences. 104 (25), 10376-10381 (2007).
- Gross, H., et al. The Genomisotopic Approach: A Systematic Method to Isolate Products of Orphan Biosynthetic Gene Clusters. Chemistry & Biology. 14 (1), 53-63 (2007).
- Spohn, M., Wohlleben, W., Stegmann, E. Elucidation of the zinc-dependent regulation in Amycolatopsis japonicum enabled the identification of the ethylenediamine-disuccinate ([S,S ]-EDDS) genes. Environmental Microbiology. 18 (4), 1249-1263 (2016).
- Thaker, M. N., Waglechner, N., Wright, G. D. Antibiotic resistance-mediated isolation of scaffold-specific natural product producers. Nature Protocols. 9 (6), 1469-1479 (2014).
- Katz, M., Hover, B. M., Brady, S. F. Culture-independent discovery of natural products from soil metagenomes. Journal of Industrial Microbiology & Biotechnology. 43, 129-141 (2016).
- Quinn, R. A., et al. Molecular Networking as a Drug Discovery, Drug Metabolism, and Precision Medicine Strategy. Trends in Pharmacological Sciences. 38 (2), 143-154 (2017).
- Yang, J. Y., et al. Molecular Networking as a Dereplication Strategy. Journal of Natural Products. 76 (9), 1686-1699 (2013).
- Lommen, A. MetAlign: Interface-Driven, Versatile Metabolomics Tool for Hyphenated Full-Scan Mass Spectrometry Data Preprocessing. Analytical Chemistry. 81 (8), 3079-3086 (2009).
- Katajamaa, M., Miettinen, J., Oresic, M. MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics. 22 (5), 634-636 (2006).
- Pluskal, T., Castillo, S., Villar-Briones, A., Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 11 (1), 395 (2010).
- Tautenhahn, R., Patti, G. J., Rinehart, D., Siuzdak, G. XCMS Online: A Web-Based Platform to Process Untargeted Metabolomic Data. Analytical Chemistry. 84 (11), 5035-5039 (2012).
- Kuhl, C., Tautenhahn, R., Böttcher, C., Larson, T. R., Neumann, S. CAMERA: An Integrated Strategy for Compound Spectra Extraction and Annotation of Liquid Chromatography/Mass Spectrometry Data Sets. Analytical Chemistry. 84 (1), 283-289 (2012).
- Katajamaa, M., Orešič, M. Data processing for mass spectrometry-based metabolomics. Journal of Chromatography A. 1158, 318-328 (2007).
- Liu, W. -. T., et al. Interpretation of Tandem Mass Spectra Obtained from Cyclic Nonribosomal Peptides. Analytical Chemistry. 81 (11), 4200-4209 (2009).
- Ng, J., et al. Dereplication and de novo sequencing of nonribosomal peptides. Nature Methods. 6 (8), 596-599 (2009).
- Liaw, C., et al. Vitroprocines, new antibiotics against Acinetobacter baumannii, discovered from marine Vibrio sp. QWI-06 using mass-spectrometry-based metabolomics approach. Scientific Reports. 5 (1), 1-11 (2015).
- Kang, K. B., et al. Targeted Isolation of Neuroprotective Dicoumaroyl Neolignans and Lignans from Sageretia theezans Using in Silico Molecular Network Annotation Propagation-Based Dereplication. Journal of Natural Products. 81 (8), 1819-1828 (2018).
- Wang, M., et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nature Biotechnology. 34 (8), 828-837 (2016).
- Doroghazi, J. R., et al. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nature Chemical Biology. 10 (11), 963-968 (2014).
- Medema, M. H., et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Research. 39, 339-346 (2011).
- Weber, T., et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Research. 43, 237-243 (2015).
- Blin, K., et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Research. 47, 81-87 (2019).
- Watrous, J., et al. Mass spectral molecular networking of living microbial colonies. Proceedings of the National Academy of Sciences. 109 (26), 1743-1752 (2012).
- Paulo, B. S., Sigrist, R., Angolini, C. F. F., De Oliveira, L. G. New Cyclodepsipeptide Derivatives Revealed by Genome Mining and Molecular Networking. ChemistrySelect. 4 (27), 7785-7790 (2019).
- Gonzaga de Oliveira, L., Sigrist, R., Sachetto Paulo, B., Samborskyy, M. Whole-Genome Sequence of the Endophytic Streptomyces sp. Strain CBMAI 2042, Isolated from Citrus sinensis. Microbiology Resource Announcements. 8 (2), 1-2 (2019).
- Aziz, R. K., et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics. 9 (1), 75 (2008).
- Nah, H. -. J., Pyeon, H. -. R., Kang, S. -. H., Choi, S. -. S., Kim, E. -. S. Cloning and Heterologous Expression of a Large-sized Natural Product Biosynthetic Gene Cluster in Streptomyces Species. Frontiers in Microbiology. 8, 1-10 (2017).
- Zhang, J. J., Tang, X., Moore, B. S. Genetic platforms for heterologous expression of microbial natural products. Natural Product Reports. 36 (9), 1313-1332 (2019).
- Alduina, R., et al. Artificial chromosome libraries of Streptomyces coelicolor A3(2) and Planobispora rosea. FEMS Microbiology Letters. 218 (1), 181-186 (2003).
- Jones, A. C., et al. Phage P1-Derived Artificial Chromosomes Facilitate Heterologous Expression of the FK506 Gene Cluster. PLoS One. 8 (7), 69319 (2013).
- Gomez-Escribano, J. P., Bibb, M. J. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microbial Biotechnology. 4 (2), 207-215 (2011).
- Cannell, R. J. P. . Natural Products Isolation. , (1998).
- Kersten, R. D., et al. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nature Chemical Biology. 7 (11), 794-802 (2011).
- Kersten, R. D., et al. Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules. Proceedings of the National Academy of Sciences. 110 (47), 4407-4416 (2013).
- Liu, W., et al. MS/MS-based networking and peptidogenomics guided genome mining revealed the stenothricin gene cluster in Streptomyces roseosporus. The Journal of Antibiotics. 67 (1), 99-104 (2014).
- Duncan, K. R., et al. Molecular Networking and Pattern-Based Genome Mining Improves Discovery of Biosynthetic Gene Clusters and their Products from Salinispora Species. Chemistry & Biology. 22 (4), 460-471 (2015).
- Cao, L., et al. MetaMiner: A Scalable Peptidogenomics Approach for Discovery of Ribosomal Peptide Natural Products with Blind Modifications from Microbial Communities. Cell Systems. , (2019).
- Chen, L. -. Y., Cui, H. -. T., Su, C., Bai, F. -. W., Zhao, X. -. Q. Analysis of the complete genome sequence of a marine-derived strain Streptomyces sp. S063 CGMCC 14582 reveals its biosynthetic potential to produce novel anti-complement agents and peptides. PeerJ. 7 (1), 6122 (2019).
- Kim Tiam, S., et al. Insights into the Diversity of Secondary Metabolites of Planktothrix Using a Biphasic Approach Combining Global Genomics and Metabolomics. Toxins. 11 (9), 498 (2019).
- Özakin, S., Ince, E. Genome and metabolome mining of marine obligate Salinispora strains to discover new natural products. Turkish Journal of Biology. 43 (1), 28-36 (2019).
- Trivella, D. B. B., de Felicio, R. The Tripod for Bacterial Natural Product Discovery: Genome Mining, Silent Pathway Induction, and Mass Spectrometry-Based Molecular Networking. mSystems. 3 (2), 00160 (2018).
- Maansson, M., et al. An Integrated Metabolomic and Genomic Mining Workflow To Uncover the Biosynthetic Potential of Bacteria. mSystems. 1 (3), 1-14 (2016).
- Blin, K., Kim, H. U., Medema, M. H., Weber, T. Recent development of antiSMASH and other computational approaches to mine secondary metabolite biosynthetic gene clusters. Briefings in Bioinformatics. 20 (4), 1103-1113 (2019).
- Fisch, K. M. Biosynthesis of natural products by microbial iterative hybrid PKS-NRPS. RSC Advances. 3 (40), 18228-18247 (2013).
- Tatsuno, S., Arakawa, K., Kinashi, H. Analysis of Modular-iterative Mixed Biosynthesis of Lankacidin by Heterologous Expression and Gene Fusion. The Journal of Antibiotics. 60 (11), 700-708 (2007).
- Helfrich, E. J. N., Piel, J. Biosynthesis of polyketides by trans-AT polyketide synthases. Natural Product Reports. 33 (2), 231-316 (2016).

