Model of Ischemic Heart Disease and Video-Based Comparison of Cardiomyocyte Contraction Using hiPSC-Derived Cardiomyocytes
Summary
We present a model of ischemic heart disease using cardiomyocytes derived from human induced pluripotent stem cells, together with a method for quantitative evaluation of tissue damage caused by ischemia. This model can provide a useful platform for drug screening and further research on ischemic heart disease.
Abstract
Ischemic heart disease is a significant cause of death worldwide. It has therefore been the subject of a tremendous amount of research, often with small-animal models such as rodents. However, the physiology of the human heart differs significantly from that of the rodent heart, underscoring the need for clinically relevant models to study heart disease. Here, we present a protocol to model ischemic heart disease using cardiomyocytes differentiated from human induced pluripotent stem cells (hiPS-CMs) and to quantify the damage and functional impairment of the ischemic cardiomyocytes. Exposure to 2% oxygen without glucose and serum increases the percentage of injured cells, which is indicated by staining of the nucleus with propidium iodide, and decreases cellular viability. These conditions also decrease the contractility of hiPS-CMs as confirmed by displacement vector field analysis of microscopic video images. This protocol may furthermore provide a convenient method for personalized drug screening by facilitating the use of hiPS cells from individual patients. Therefore, this model of ischemic heart disease, based on iPS-CMs of human origin, can provide a useful platform for drug screening and further research on ischemic heart disease.
Introduction
Ischemic heart disease (IHD) is recognized worldwide as the leading cause of death, and it was estimated to be responsible for more than nine million fatalities in 20161. The prevalence of cardiovascular disease continues to rise, and globalization seems to have contributed to the prevalence of heart disease risk factors in developing countries. Therefore, the study of IHD is becoming increasingly more urgent2.
Experimental models of cardiovascular disease are critical for studying the mechanisms of disease, accuracy of diagnosis, and development of new therapies. Several experimental models have been proposed by many laboratories. It is of utmost importance to choose a model with significant advantages; feasibility, repeatability and similarity to human disease are key factors in the selection of cardiovascular disease models3. Human induced pluripotent stem cells (hiPSCs) carrying specific cardiomyopathy-associated mutations are a promising alternative to animal models4. Although many strategies have been described for inducing cardiomyocytes using iPSCs5,6,7,8,9, here we provide a simple method to produce an IHD model using cardiomyocytes differentiated from hiPSCs, in which laborious selection of cardiomyocyte using markers is not applied. In this method, cardiomyocytes are selected functionally by analyzing spontaneous contraction.
Induction of cardiomyocytes using hiPSCs avoids animal sacrifice and technically difficult surgery. Establishing animal models of IHD requires challenging surgical techniques10. Perfectly simulating the various pathophysiological aspects of human heart disease such as heart rate and action potentials from the physiology of rodents is nearly impossible. Coupled with the morality and ethics of using animal models, the development of new experimental models other than animal models is imperative. Cardiomyocytes differentiated from human iPS cells better mimic the physiological state of the human heart. In this protocol, we establish a model of IHD using cardiomyocytes derived from hiPS cells (hiPS-CMs). In our model, the deprivation of oxygen and glucose leads to a decrease in the contractile force and viability of hiPS-CMs. Our method provides a new approach to model IHD and demonstrates a new platform for the study of this disease.
Protocol
1. hiPSC maintenance culture
- Coat a six-well culture plate with laminin (Table of Materials).
- Dilute laminin to 0.5 μg/mL in phosphate buffered saline (PBS).
- Add diluted laminin to a six-well plate at a volume of 2.0 mL/well.
- Incubate the plate at 37 °C for 30 min.
- Remove the laminin solution from the wells.
- Seed hiPSCs in 2 mL of iPSC maintenance medium (Table of Materials) onto the coated wells at a density of 3 × 104 cells/well without drying the surface.
- Subculture the hiPSCs.
- Prepare 0.5× cell dissociation enzymes (Table of Materials) solution.
- Mix 10 μL of 0.5 M Ethylenediaminetetraacetic (EDTA) and 10 mL of PBS to make 0.5 mM EDTA/PBS.
- Dilute 10 mL of 1× cell dissociation enzymes to 0.5× by adding 10 mL of 0.5 mM EDTA/PBS.
- Aspirate the spent medium from the wells.
- Add 2 mL/well of PBS to wash the cells, then discard the PBS.
- Add 800 μL of the 0.5× cell dissociation enzymes, and incubate the cells for 7 min at 37 °C in a humidified incubator containing 5% CO2.
NOTE: 0.05% trypsin-EDTA can be used to dissociate cells. - Gently remove the 0.5× cell dissociation enzymes solution.
- Wash the cells with 2 mL PBS, then discard the PBS.
NOTE: Be gentle because the cells easily detach after adding cell dissociation enzymes solution. - Add 1 mL of iPS maintenance medium (Table of Materials) containing 10 μM Y-27632.
NOTE: Adding Y-27632 increases the survival rate of hiPSCs. - Dislodge cells using a cell scraper, and collect them in a 15 mL centrifuge tube.
- Count the number of cells. Adjust the cell density to 1.5 × 104 cells/mL by adding iPS maintenance medium containing 10 μM Y-27632.
NOTE: Do not use centrifugation in order to prevent cell damage. - Seed 2 mL of cell mixture on the laminin-coated six-well plate (final density: 3 × 104 cells/well).
- Incubate the cells at 37 °C in a humidified incubator containing 5% CO2.
- Replace the culture medium with iPS maintenance medium without Y-27632 on days 1, 4, 5, and 6.
- Prepare 0.5× cell dissociation enzymes (Table of Materials) solution.
- Subculture the cells on day 7.
NOTE: Subculture the cells by day 7 in order to inhibit random differentiation of hiPSCs.
2. Induction of cardiac differentiation of hiPSCs
- Coat a 96-well culture plate with laminin (Table of Materials).
- Dilute laminin to 1.675 μg/mL with PBS.
- Add the diluted laminin solution to a 96-well plate at a volume of 0.1 mL/well.
- Incubate the plate at 37 °C for 30 min.
- Seed hiPSCs onto a 96-well plate at a density of 3 × 104 cells/well.
- Proliferate hiPSCs at 37 °C in a humidified incubator containing 5% CO2.
- One day later, replace the medium with 200 μL/well iPS growth medium (Table of Materials).
- Change the medium every day for an additional 2–3 days until the cells reach 70%–80% confluency.
- Apply differentiation media.
- Aspirate the spent medium and replace it slowly with 200 μL/well of pre-warmed differentiation medium A (Table of Materials).
- Place the plate at 37 °C in a humidified incubator containing 5% CO2.
- After 48 h, aspirate the medium and slowly replace it with 200 μL/well of pre-warmed differentiation medium B (Table of Materials).
- Place the plate at 37 °C in a humidified incubator containing 5% CO2.
NOTE: It is extremely important that the differentiation media are kept fresh. They will gradually lose their differentiation effect, typically within two weeks. If necessary, aliquot fresh medium and store at −20 °C.
- Apply cardiomyocyte maintenance medium.
- After 48 h, aspirate the medium and slowly replace it with 200 μL/well of pre-warmed cardiomyocyte maintenance medium (Table of Materials).
- Place the plate at 37 °C in a humidified incubator containing 5% CO2.
- Replace the cardiomyocyte differentiation medium every other day up to day 30.
NOTE: It is important that the duration of the application of differentiation media A and B is accurately set to 48 h to ensure the precise expression sequence of genes required for differentiation.
3. Exposure to ischemia
- Deprive the culture medium of nutrients and oxygen.
- Prepare Dulbecco's modified Eagle medium (DMEM) without glucose and serum.
- Aspirate culture medium from the wells of the 96-well plate containing hiPS-CMs.
- Add the nutrient-deprived medium to the wells at a volume of 200 μL/well.
- Place the culture plate in a hypoxic incubator (Table of Materials).
- Infuse nitrogen gas, and maintain the internal oxygen concentration at 2% and CO2 concentration at 5% for 24 h.
- Proceed to desired analyses: e.g., viability assay, assessment of contractility, or assessment of cellular damage.
4. Assessment of cellular viability using MTT assay
- Use the MTT assay kit (Table of Materials) to quantitatively assess the viability of cells.
NOTE: Exposure to 1 mM hydrogen peroxide in DMEM for 1 h can be used to damage cells (positive control). Exposure to 0 mM hydrogen peroxide in DMEM can be used for the negative control.- Add 10 μL of MTT reagent to the cells using a repeating pipettor.
- Mix gently for one minute on an orbital shaker.
- Incubate the cells for 3–4 h at 37 °C in a 5% CO2 incubator. After incubation, the formazan produced in the cells will appear as dark crystals in the bottom of the wells.
- After removing the supernatant, dissolve the insoluble formazan crystals in 100 μL of dimethyl sulfoxide solution (DMSO). This solution will dissolve the formazan crystals, producing a purple solution.
CAUTION: DMSO can irritate eyes, respiratory system, and skin. Wear suitable gloves and eye/face protection. - Measure the absorbance of each sample with a microplate reader at a wavelength of 570 nm.
5. Assessment of contractility of iPS-CMs
- If not installed, obtain Particle Image Velocimetry ImageJ plugin11 at https://sites.google.com/site/qingzongtseng/piv and install.
- Using a phase contrast microscope, record the video images of hiPS-CMs using the 4× objective lens at ~20 frames per second for ~10 s, and save as “analyze.avi”. For a comparison of contractility between before and after ischemia, confirm that the location of interest is recorded.
NOTE: A microscope with an automated stage is useful to target the location of interest. Movie file should be in .avi format for later analysis. If not, convert the movie to .avi. - Create folder structure as indicated in Figure 1. An example of “joblist.txt” is indicated in supplementary file.
- Analyze discrete two-dimensional vector fields of cellular displacement.
- Launch Fiji (ImageJ) software12, go to Plugins > Macros > Edit and open “vector_analysis.ijm” (Supplementary Coding File).
- Click Run. Analysis will be performed automatically.
NOTE: Displacement vectors D(x,y) are calculated for every 16 × 16 pixels between the reference frame (the first frame) and all subsequent frames (frames 1 vs. 2, 1 vs. 3, 1 vs. 4, etc.). The result is calculated for every frame and saved as “vec_x.txt” (x is a frame number). A vector of maximum displacement, M(x, y), is defined for every (x, y) pair as follows: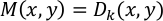
where k represents the frame number at which |Dk (x,y)| = max [|D2 (x,y)|, |D3 (x,y)|, …, |Dn (x,y)|] and n denotes the last frame. The result is saved as “Max_vector.txt”. |M(x, y)| represents the maximum displacement caused by cardiomyocyte contraction at the analysis point (x, y). Contractility value C in arbitrary units is calculated as follows: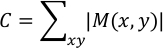
This value is saved at column 7, row 1 in the “Max_vector.txt”. C represents the sum of displacement for all (x, y). The larger the portion where the displacement due to myocardial contraction is large, the larger the value of C. The vector field of maximum displacement, M(x, y), is overlaid with phase contrast image of the first frame (“Phase_contrast.png”) and saved as “Overlaid.png”. As the magnitude of the displacement vector is calculated based on the first frame of the video, it is preferable for the hiPS-CMs to be at resting diastolic period for the first frame.
6. Assessment of cellular damage using flow cytometry
- Dilute a 1 mg/mL solution of propidium iodide to 1:1,000 in PBS.
- Stain the detached cells with propidium iodide.
- Aspirate the medium and place it in an appropriately sized centrifuge tube.
- Centrifuge the tube at 1,000 × g for 5 min, and carefully remove the supernatant so as not to lose sedimented cells.
- Incubate the cells with the diluted propidium iodide solution at room temperature for 15 min in the dark.
- Centrifuge the tube at 1,000 × g for 5 min, and carefully remove the supernatant so as not to lose sedimented cells.
- Reconstitute the cells in ~1 mL of PBS.
NOTE: It is important to collect detached floating cells from the medium to accurately quantify cellular damage.
- Stain attached cells with propidium iodide.
- Wash attached cells twice with PBS gently, then discard PBS.
- Incubate the cells with the diluted propidium iodide solution for 15 min in the dark.
- Aspirate the propidium iodide solution.
- Detach the cells using 0.25% trypsin. Move the cell solution into a centrifuge tube.
- Centrifuge the tube at 1,000 × g for 5 min, and carefully remove the supernatant so as not to lose sedimented cells.
- Reconstitute the cells in ~1 mL of PBS.
- Mix the floating and attached cells for fluorescence activated cell sorting (FACS) analysis.
- Pass the cell solution through a 30 μm filter.
NOTE: Passing the cells to a filter is very important for an accurate FACS measurement. - Analyze the samples using a FACS system.
7. Immunostaining
- Fix the cell sample.
- Aspirate the culture medium.
- Add 4% paraformaldehyde in PBS for 10 min at room temperature.
- Wash the cells three times with PBS.
NOTE: Fresh paraformaldehyde solution is recommended for optimal fixation.
- Permeabilize the cells with 0.2% Triton X-100 for 15 min, then discard the reagent.
- Add 3% bovine serum albumin to block the cells for 30 min.
- Apply primary antibody.
- Discard bovine serum albumin solution from the culture plate.
- Incubate the cells with primary antibodies overnight at 4 °C.
- Remove the antibody solution.
- Wash the cells three times with PBS.
- Apply secondary antibody.
- Discard PBS solution from the culture plate.
- Incubate the cells with secondary antibodies for 30 min at room temperature in the dark.
NOTE: Primary anti-cardiac troponin T (TNNT2) mouse monoclonal antibody is used at a dilution of 1:750 in 3% BSA. The Alexa Fluor 488-conjugated secondary goat anti-mouse antibody is diluted to 1:1,000 in 3% BSA.
- Additional staining of nucleus and actin fibers.
- Remove the antibody solution.
- Incubate the cells in nucleus staining reagent (Table of Materials) and actin staining reagent (Table of Materials) in PBS for 30 min at room temperature in the dark.
NOTE: 4’, 6-diamidino-2-phenylindole (DAPI) or Hoechst 33342 can be used for nuclear staining. - Wash the cells three times with PBS.
- Capture fluorescence images using a fluorescence microscope.
Representative Results
Successfully differentiated cells show spontaneous contraction under the microscope (Video 1). Typically, 50% of wells show spontaneous contraction in <20 days (Supplementary Figure 1). Cardiac marker protein (e.g., cTnT) can be used to confirm successful differentiation (Figure 2).
Typically, cells from the ischemic group show lower viability in MTT assays (Figure 3A) and contractility (Figure 3E,F, Supplementary Figure 2) than those from normoxic control group. Also, the ratio of propidium iodide-positive cells is higher in the ischemic group than in the control group (Figure 3B–D), which indicates higher cellular damage.
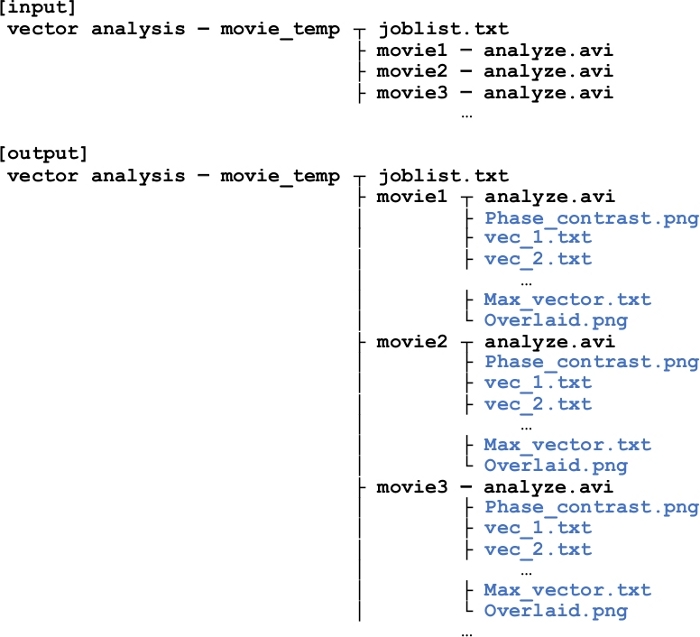
Figure 1: Folder structure for displacement vector analysis using imageJ.
“joblist.txt” describes path to movie files each line. If there are three files to analyze, there will be three folders (movie1, movie2, and movie3), in which “analyze.avi” (the movie of interest) should be placed. After the analysis is performed by “vector_analysis.ijm” code, files will be generated (indicated in blue) in each movie folder. Information stored in each file is explained in detail in the main text. Please click here to view a larger version of this figure.
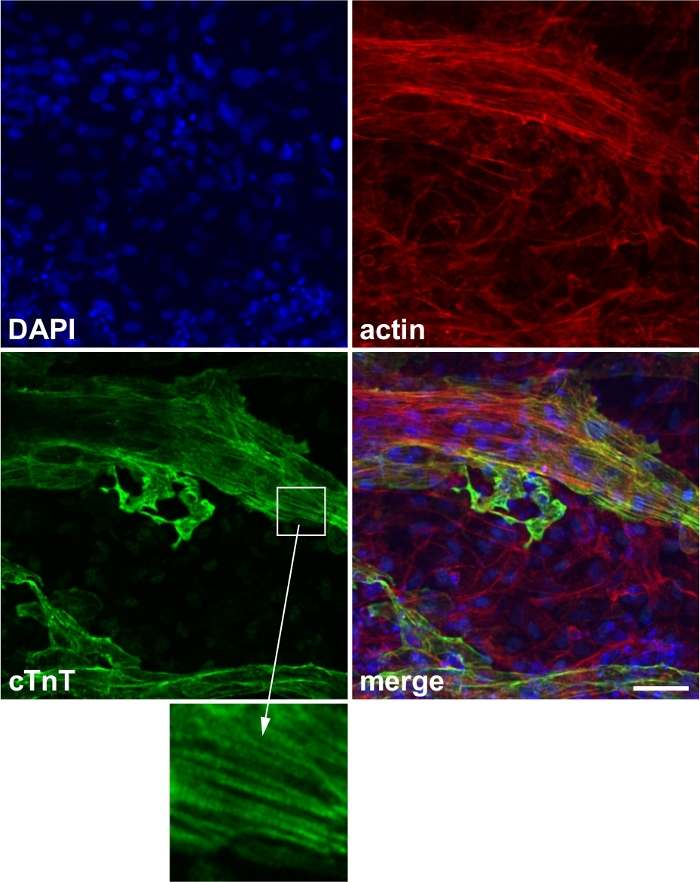
Figure 2: Representative image of cardiac marker staining.
Expression of the cardiac marker protein cardiac troponin T (cTnT). Blue: 4’,6-diamidino-2-phenylindole (DAPI), red: actin, green: cTnT. Inset: striated expression of cTnT, which corresponds to the sarcomere structure. Scale bar, 50 μm. This figure has been modified from Wei et al.13. Please click here to view a larger version of this figure.
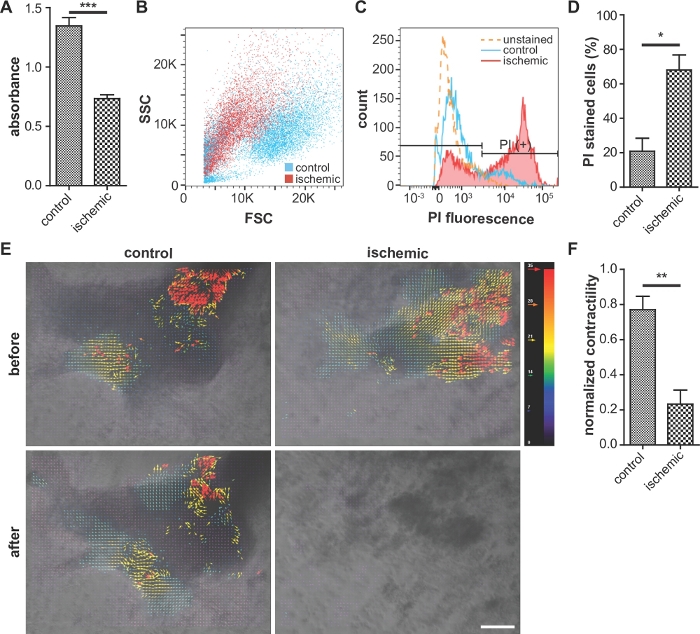
Figure 3: Representative images of cellular viability assay, assessment of cellular damage, and contractility.
(A) Comparison of cellular viability (MTT assay). Higher absorbance indicates higher viability. n = 5 for each condition. (B, C, and D) Flow cytometry analysis of propidium iodide (PI)-stained cells. Ischemic cells exhibit a reduced forward scatter intensity and increased PI fluorescence, compared with the control. (D) Percentage of PI-stained cells in the flow cytometry analysis. n = 3 for each condition. (E) Analysis of iPS-CMs contractility using ImageJ software. The red and blue vectors indicate the largest and smallest contractions, respectively. Scale bar, 100 μm. (F) Quantitative analysis of the iPS-CM contractility using code. n = 3 for control and n = 8 for ischemic condition. Error bars represent standard error of the mean. For the statistical analysis, unpaired two-tailed Student’s t-test was performed. *: p < 0.05, **: p < 0.01. ***: p < 0.0001. This figure is cited from Wei et al.13 Please click here to view a larger version of this figure.
Video 1: Differentiated cells show spontaneous contraction under the microscope. Please click here to download this video.
Supplementary Figure 1: Kaplan-Meier analysis of percentage of samples without spontaneous contraction. 50% of iPS-CM samples showed contraction on day 20. On day 30, 64.4% of the samples showed contraction. n = 83. Please click here to download this figure.
Supplementary Figure 2: Assessment of cardiac contractility. Scaled contractility obtained by dividing contractility C by the number of analysis points (x, y) was plotted for control and ischemic groups before and after the exposure to 24 h hypoxia. n = 3 for control and n = 8 for ischemic condition. Please click here to download this figure.
Supplementary Figure 3: Assessment of the content of cardiomyocytes. Immunostaining of cardiac marker protein on a 96 well plate. Green: cTnT, Blue: DAPI. Rate of the differentiated cells was calculated to be 20.7 ± 9.6% by dividing the area of cTnT-stained region by that of DAPI-stained region. Scale bar = 1 mm. n = 4 for control condition. Please click here to download this figure.
Supplementary Coding File. Please click here to download this file.
Discussion
Researchers often use laboratory small-animal models to conduct IHD experiments. Here, we developed a cell culture model of human IHD to conduct such experiments.
The main problem a user of this protocol may face is low rate of differentiated cardiomyocytes. Several steps should be taken with great care to improve the rate of successful differentiation: (a) be gentle when pipetting because the cells easily detach after addition of the cell dissociation enzymes reagent, (b) adding Y-27632 increases the survival rate of iPS cells when detaching, and (c) the timing of medium changes at the beginning of differentiation should be precisely 48 h apart to ensure controlled expression of genes involved in differentiation.
Regarding the extracellular matrix proteins used for coating the surface of culture plate, other materials than laminin we used in this protocol can be used. For example, Matrigel14,15, and gelatin16,17 are used for the feeder-free maintenance culture of hiPSCs. According to Haraguchi et al., hiPSCs seeded on Matrigel were successfully differentiated into cardiac cell sheet18.
There are a number of preceding studies regarding culture models for disease modeling using cardiomyocytes derived from hiPSCs. Regarding the modeling of ischemia-reperfusion injury, manipulations of the cellular environment, such as hyperkalemia, acidosis, and lactate accumulation, have been introduced19. Other methods include cell pelleting to hinder oxygen diffusion20 and metabolic inhibition using cyanide21. In the current protocol, cell injury was achieved by relatively simple method, namely 24 h deprivation of oxygen and nutrients. However, care should be taken to consider accurate pathophysiological processes of the ischemic heart disease, as there are indeed differences between the disease in vivo and the disease model of the current protocol, such as the presence or absence of three-dimensional cellular environment and blood.
Regarding the technological aspect of the evaluation of cardiomyocyte function, Toepfer et al. reported a MatLab-based algorithm to determine sarcomere contraction and relaxation in hiPS-CMs22. Smith et al. reported an advanced method of high-throughput electrophysiological analysis of excitable cells derived from hiPS-CMs, using sophisticated nanotopographically patterned multielectrode arrays23,24. The advantage of our protocol is that it only requires conventional software and consumables, such as imageJ software and 96-well plates.
With respect to the oxygen level in the heart, the oxygen pressure in veins that reach the heart is thought to be 40 mmHg25. According to McDougal et al., the extracellular oxygen pressure under hypoxia is estimated to be <12.8 mmHg26. By applying the method of Rounds et al.27, the oxygen pressure in the culture medium (salinity: 35‰) under the hypoxic condition (2% oxygen) at 37 °C treated with the current protocol is calculated to be 14.9 mmHg, which is higher than the estimation above. Interestingly, Al-Ani et al. reported that there is a gradient of oxygen pressure in the culture medium, and the oxygen pressure is affected by cell type, seeding density, and medium volume28. Typically, the oxygen concentration at the bottom of the culture plate where cells reside is the lowest. Therefore, the effect of depth in the culture medium would further reduce the effective oxygen pressure near hiPS-CMs. In order to induce sufficient damage to hiPS-CMs using hypoxic condition, careful attention must be paid to the depth of medium and the density of cells.
Our hiPS-CM model, which is very close to the physiological conditions of human cardiac myocytes, advantageously mimics human IHD. Animal model-based approaches include ethical, technical, and academic issues. Particularly, in vivo models require an advanced technique of microsurgery to achieve reproducible data: e.g., occlusion of the anterior descending branch of the left coronary artery in rodents3. The hiPS-CM model described herein overcomes these critical barriers and provides a useful, relevant, and repeatable platform for cardiovascular disease.
However, some limitations should be noted. An obvious difference between iPS-induced cardiomyocytes and normal cardiomyocytes is the absence of T-tubules29, and we did not include humoral factors such as tissue damage induced by leukocytes and activation of a complement system in our study. Moreover, the rate of differentiated cardiomyocytes in this model (20.7 ± 9.6%, Supplementary Figure 3) should be improved. The recent publication by Halloin et al. describes a scalable, chemically defined method to induce hiPS-CM purity of >95% by chemical WNT pathway modulators without requirement of any selection by markers30. Nevertheless, our model of human IHD is relatively simple and clinically applicable (e.g., drug screening using patient-derived iPS cells). Our model is also a unique platform to further elucidate mechanisms underlying IHDs.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This study was supported by JSPS KAKENHI, Fund for the Promotion of Joint International Research (Fostering Joint International Research), 17KK0168. The authors gratefully acknowledge Central Research Laboratory, Okayama University Medical School for the assistance of FACS.
Materials
| Actin staining reagent (ActinRed 555 ReadyProbes Reagent) | Thermo Fisher Scientific, Waltham, MA, USA | R37112 | |
| Anti cardiac troponin T antibody | Thermo Fisher Scientific, Waltham, MA, USA | MA5-12960 | |
| Cardiomyocyte maintenance medium (PSC Cardiomyocyte Differentiation Kit) | Thermo Fisher Scientific, Waltham, MA, USA | A2921201 | |
| Cell dissociation enzymes (TrypLE Select) | Thermo Fisher Scientific, Waltham, MA, USA | 12563011 | |
| Differentiation medium A (PSC Cardiomyocyte Differentiation Kit) | Thermo Fisher Scientific, Waltham, MA, USA | A2921201 | |
| Differentiation medium B (PSC Cardiomyocyte Differentiation Kit) | Thermo Fisher Scientific, Waltham, MA, USA | A2921201 | |
| FACS Aria III system | BD Biosciences, San Jose, CA, USA | ||
| Fluorescenct microscope | KEYENCE, Osaka, Japan | BZ-X710 | |
| Goat anti-mouse Alexa Fluor 488 secondary antibody | Thermo Fisher Scientific, Waltham, MA, USA | A28175 | |
| Human iPS cells | RIKEN, Tsukuba, Japan | 201B7 | |
| Hypoxic incubator | SANYO, Osaka, Japan | MCO-175M | |
| iPSC growth medium (Essential 8 Medium) | Thermo Fisher Scientific, Waltham, MA, USA | A1517001 | |
| iPSC maintenance medium (StemFit AK02N) | Ajinomoto, Tokyo, Japan | RCAK02N | |
| Laminin (iMatrix-511) | Nippi, Tokyo, Japan | iMatrix-511 | |
| MTT assay kit (MTT Cell Proliferation Assay Kit) | Cayman Chemical Company, Ann Arbor, MI, USA | 10009365 | |
| Nucleus staining reagent (NucBlue Fixed Cell ReadyProbes Reagent) | Thermo Fisher Scientific, Waltham, MA, USA | R37606 | |
| Triton X-100 | Nacalai Tesque, Kyoto, Japan | 35501-02 |
References
- Naghavi, M. Global Burden of Disease Self-Harm, C. Global, regional, and national burden of suicide mortality 1990 to 2016: systematic analysis for the Global Burden of Disease Study 2016. BMJ. 364, 94 (2019).
- Nowbar, A. N., Gitto, M., Howard, J. P., Francis, D. P., Al-Lamee, R. Mortality From Ischemic Heart Disease. Circulation: Cardiovascular Quality and Outcomes. 12 (6), 005375 (2019).
- Oh, J. G., Ishikawa, K. Experimental Models of Cardiovascular Diseases: Overview. Methods in Molecular Biology. 1816, 3-14 (2018).
- Brodehl, A., et al. Human Induced Pluripotent Stem-Cell-Derived Cardiomyocytes as Models for Genetic Cardiomyopathies. International Journal of Molecular Sciences. 20 (18), (2019).
- Garbern, J. C., et al. Inhibition of mTOR Signaling Enhances Maturation of Cardiomyocytes Derived from Human Induced Pluripotent Stem Cells via p53-Induced Quiescence. Circulation. , (2019).
- Horikoshi, Y., et al. Fatty Acid-Treated Induced Pluripotent Stem Cell-Derived Human Cardiomyocytes Exhibit Adult Cardiomyocyte-Like Energy Metabolism Phenotypes. Cells. 8 (9), (2019).
- Hu, D., et al. Metabolic Maturation of Human Pluripotent Stem Cell-Derived Cardiomyocytes by Inhibition of HIF1alpha and LDHA. Circulation Research. 123 (9), 1066-1079 (2018).
- Correia, C., et al. Distinct carbon sources affect structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Scientific Reports. 7 (1), 8590 (2017).
- Yang, X., et al. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. Journal of Molecular and Cellular Cardiology. 72, 296-304 (2014).
- Tamargo, J., et al. Genetically engineered mice as a model for studying cardiac arrhythmias. Frontiers in Bioscience. 12, 22-38 (2007).
- Tseng, Q., et al. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proceedings of the National Academy of Sciences of the United States of America. 109 (5), 1506-1511 (2012).
- Schneider, C. A., Rasband, W. S., Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 9 (7), 671-675 (2012).
- Wei, H., Wang, C., Guo, R., Takahashi, K., Naruse, K. Development of a model of ischemic heart disease using cardiomyocytes differentiated from human induced pluripotent stem cells. Biochemical and Biophysical Research Communications. 520 (3), 600-605 (2019).
- Ghaedi, M., Niklason, L. E. Human Pluripotent Stem Cells (iPSC) Generation, Culture, and Differentiation to Lung Progenitor Cells. Methods in Molecular Biology. 1576, 55-92 (2019).
- Hou, J., et al. Retaining mTeSR1 Medium during Hepatic Differentiation Facilitates Hepatocyte-Like Cell Survival by Decreasing Apoptosis. Cellular Physiology and Biochemistry. 51 (4), 1533-1543 (2018).
- Yoshida, K., et al. Differentiation of mouse iPS cells into ameloblast-like cells in cultures using medium conditioned by epithelial cell rests of Malassez and gelatin-coated dishes. Medical Molecular Morphology. 48 (3), 138-145 (2015).
- Guzzo, R. M., Drissi, H. Differentiation of Human Induced Pluripotent Stem Cells to Chondrocytes. Methods in Molecular Biology. 1340, 79-95 (2015).
- Haraguchi, Y., Matsuura, K., Shimizu, T., Yamato, M., Okano, T. Simple suspension culture system of human iPS cells maintaining their pluripotency for cardiac cell sheet engineering. Journal of Tissue Engineering and Regenerative. 9 (12), 1363-1375 (2015).
- Chen, T., Vunjak-Novakovic, G. In vitro Models of Ischemia-Reperfusion Injury. Regenerative Engineering and Translational Medicine. 4 (3), 142-153 (2018).
- Strijdom, H., Genade, S., Lochner, A. Nitric Oxide synthase (NOS) does not contribute to simulated ischaemic preconditioning in an isolated rat cardiomyocyte model. Cardiovascular Drugs and Therapy. 18 (2), 99-112 (2004).
- Cavalheiro, R. A., et al. Potent cardioprotective effect of the 4-anilinoquinazoline derivative PD153035: involvement of mitochondrial K(ATP) channel activation. PLoS One. 5 (5), 10666 (2010).
- Toepfer, C. N., et al. SarcTrack. Circulation Research. 124 (8), 1172-1183 (2019).
- Smith, A. S. T., et al. NanoMEA: A Tool for High-Throughput, Electrophysiological Phenotyping of Patterned Excitable Cells. Nano Letters. , (2019).
- Smith, A. S., Macadangdang, J., Leung, W., Laflamme, M. A., Kim, D. H. Human iPSC-derived cardiomyocytes and tissue engineering strategies for disease modeling and drug screening. Biotechnology Advances. 35 (1), 77-94 (2017).
- Sitkovsky, M., Lukashev, D. Regulation of immune cells by local-tissue oxygen tension: HIF1 alpha and adenosine receptors. Nature Reviews Immunology. 5 (9), 712-721 (2005).
- McDougal, A. D., Dewey, C. F. Modeling oxygen requirements in ischemic cardiomyocytes. Journal of Biological Chemistry. 292 (28), 11760-11776 (2017).
- Rounds, S. A., Wilde, F. D., Ritz, G. F. Chapter A6. Section 6.2. Dissolved oxygen. Report No. 09-A6.2. , (2006).
- Al-Ani, A., et al. Oxygenation in cell culture: Critical parameters for reproducibility are routinely not reported. PLoS One. 13 (10), 0204269 (2018).
- Cadet, J. S., Kamp, T. J. A Recipe for T-Tubules in Human iPS Cell-Derived Cardiomyocytes. Circulation Research. 121 (12), 1294-1295 (2017).
- Halloin, C., et al. Continuous WNT Control Enables Advanced hPSC Cardiac Processing and Prognostic Surface Marker Identification in Chemically Defined Suspension Culture. Stem Cell Reports. 13 (2), 366-379 (2019).

