A Triple Culture Cell System Modeling the Human Blood-Brain Barrier
Summary
This protocol describes a method to establish a human blood-brain barrier (BBB) in vitro model. The endothelial cells and pericytes are seeded on each side of an insert filter (blood compartment), and astrocytes are seeded in the bottom well (brain compartment). The model characterized was used for nanoparticles transport experiments.
Abstract
The delivery of drugs to the brain remains a challenge due to the blood-brain barrier’s (BBB) highly specific and restrictive properties, which controls and restrict access to the brain parenchyma. However, with the development of nanotechnologies, large panels of new nanomaterials were developed to improve drug delivery, highlighting the need for reliable in vitro microsystems to predict brain penetration in the frame of preclinical assays. Here is a straightforward method to set up a microphysiological system to model the BBB using solely human cells. In its configuration, the model consists of a triple culture including brain-like endothelial cells (BLECs), pericytes, and astrocytes, the three main BBB cellular actors necessary to induce and regulate the BBB properties in a more physiological manner without the requirement of tightening compounds. The model developed in a 12-well plate format, ready after 6 days of triple culture, is characterized in physical properties, gene, and protein expressions and used for polymeric nanogel transport measurement. The model can be used for an extensive range of experiments in healthy and pathological conditions and represents a valuable tool for preclinical assessments of molecule and particle transport, as well as inter-and intracellular trafficking.
Introduction
The BBB, localized at the level of brain capillary endothelial cells (ECs), controls and regulates access to the brain parenchyma, which is crucial for maintaining brain homeostasis and the function of neural cells1,2. However, in the case of brain pathology, the lack of access to the brain parenchyma represents a real obstacle to developing therapeutic strategies.
The BBB ECs possess a complex set of properties, including tight junction (TJ) proteins, which seal the intercellular space, associated with a system of efflux pumps, specific transporters, and receptors, which control the transcellular pathway1,2,3. Moreover, all these properties are induced and maintained, thanks to communications with the pericytes embedded in the BBB EC basement membrane and the astrocytes, whose end-feet surround the brain capillaries1,2,3. Hence, studying the BBB in vitro is a challenge considering the complexity of its architecture and the communications among the different cell types constituting the neurovascular unit (NVU)2. Moreover, the different cell types are crucial for the induction and maintenance of BBB properties and consequently impact the prediction of the crossing through the BBB. Different strategies for drug delivery to the brain were already tested using a large panel of tactics to bypass the BBB restricted properties4. More recently, with the progress of nanotechnologies, new materials are being developed for applications as drug carriers5,6. In addition to their higher load, reduced toxicity, and increased drugs' bioavailability, these new nanomaterials can be functionalized for a Trojan horse strategy to cross the BBB and specifically target cells in the parenchyma5,6. Among the different types of nanomaterials being evaluated, nanogels have attracted considerable attention, mainly due to their colloidal properties and ability to tailor the chemical structure to introduce stimuli-responsive properties7,8,9,10,11,12,13,14,15.
In vitro models are now developed for preclinical studies using human cells to predict brain penetration of drugs16. Different settings of these models are available, from monolayers of brain ECs to multiple cell systems16. Considering the importance of the NVU cells in the BBB induction and maintenance and the coordinated response to the pathological environment, BBB in vitro models need to consider all these protagonists to improve the relevance of the prediction2,17.
The current method describes setting up a triple culture in vitro model of the human BBB, which is fully developed with human cells to study specific cellular and human molecular mechanisms. To be physiologically relevant, the model consists of the main three cellular actors of the BBB (ECs, pericytes, and astrocytes) necessary to induce and maintain the BBB properties, without the use of tightening compounds and displaying a set of properties required to be considered as an in vitro BBB model16,18. The model is set up in a configuration delimiting the blood and brain compartment, suitable for preclinical studies of drug and particle transport to predict brain penetration. The usefulness of the model is illustrated by measuring the transport of polymeric nanogels.
Protocol
The protocol was approved by the French Ministry of Higher Education and Research (reference: CODECOH DC2011-1321) and by the local investigational review board (Béthune Maternity Hospital, Beuvry, France). For obtaining the endothelial cells (ECs), written and informed consent from the donor's parents was obtained to collect umbilical cord blood, in compliance with the French Legislation. The pericytes are provided by Professor Takashi Kanda (Department of Neurology and Clinical Neuroscience, Yamaguchi University Graduate School of Medicine, Ube, Japan) that were isolated as per Reference19. Primary human brain cortex astrocytes are purchased from a commercial provider (see Table of Materials).
1. Cell culture
- Culturing of endothelial cells
NOTE: Endothelial cells (ECs) are derived from CD34+ hematopoietic stem cells isolated from human umbilical cord blood, according to the method described by Pedroso et al.20. The endothelial phenotype of the ECs is described in Pedroso et al.20.- Cultivate human ECs using endothelial cell medium supplemented with 5% of fetal calf serum (FCS), 0.5% of gentamicin, and 1% of endothelial cell growth supplement (ECM) (see Table of Materials).
- For sub-culturing ECs, two days before the setting of the model, coat one dish with 10 mL of 2% gelatin for 15 min at 37 °C and then replace it with 20 mL of warm ECM. Thaw one vial of ECs containing 1 million cells (cells were manually counted as described in step 2.3.3 before cell freezing) in the pre-coated cell culture dish.
- After 3 h at 37 °C, renew the medium and maintain the cells until the setting of the triculture in a humidified atmosphere inside an incubator at 37 °C under 5% CO2 and 21% O2.
- Culturing of pericytes
NOTE: The pericytes are isolated from the human brain according to the protocol published by Shimizu et al.19, whose isolation procedure follows the method published by Kanda et al.21 with modifications.- Cultivate the pericytes using Dulbecco's modified Eagle medium supplemented with 4.5 g/L of glucose (DMEM HG), 10% of FCS, 1% de penicillin/streptomycin, and 1% of L-glutamine (see Table of Materials).
- For pericyte sub-culturing, five days before the setting of the model, coat two dishes with 8 mL/dish of 100 µg/mL of collagen type I solution in 0.02 N acetic acid for 1 h at room temperature (RT) and then wash twice with RT DMEM HG. Thaw one vial of pericytes containing 1 million cells in a conical tube containing 10 mL of warm medium and centrifuge the suspension for 5 min at 190 x g at 20 °C.
- Resuspend the pellet in 10 mL of warm medium and seed in the pre-coated cell culture dishes prefilled with 15 mL of warm medium/dish. The medium is renewed after 3 days, and the cells are maintained until the setting of the triculture in a humidified incubator at 37 °C under 5% CO2 and 21% O2.
- Culturing of astrocytes
- Cultivate the astrocytes using a astrocyte medium supplemented with 20% of FCS, 1% of astrocyte growth supplement, and 1% of penicillin/streptomycin solution (AM) (see Table of Materials).
- For sub-culturing the astrocytes, one week before the setting of the model, coat one T75 cell culture flask with 10 mL of 2 µg/cm2 poly-L-lysine (PLL) for 1 h at 37 °C and wash twice with RT sterile water. Thaw one vial of astrocytes containing 1 million cells in 20 mL of warm medium and seed in the pre-coated T75 cell culture flask.
NOTE: The commercially obtained cell vials confirm the presence of ~1 million cells, so the counting of cells was not performed here. - Maintain the cells in a humidified incubator at 37 °C under 5% CO2 and 21% O2. The medium is renewed after 24 h and then every 2 days until the setting of the triculture.
2. Triple culture model setting
NOTE: The assembly of the three cell types is performed on the same day. The day before the setting of the triculture, perform the collagen type I coating on the reverted insert filters (see Table of Materials) and seed the astrocytes in the PLL-precoated wells of a 12-well plate.
- Seeding of astrocytes in the wells
- Coat the wells with 500 µL of 2 µg/cm2 PLL solution as described in step 1.3.2.
- Wash the cells once with 10 mL of warm phosphate buffer saline – calcium and magnesium-free 1X (1X PBS-CMF) (Table 1) before incubating for 3 min at 37 °C with 10 mL of warm 20% trypsin/EDTA (T/E) solution and mechanically detach the cells from the flask. Transfer the suspension to a conical tube containing 5 mL of warm non-diluted FCS.
NOTE: According to the provider's protocol22, the collection of astrocytes can be optimized by placing the flask in the incubator for 1 min and tapping the flask to help complete the detachment. The remaining cells should be collected with 5 mL of T/E neutralization solution and placed in the FCS-containing conical tube. - Centrifuge the suspension for 5 min at 20 °C at 190 x g.
- Resuspend the cell pellet in 5 mL of warm AM. Count the cells by diluting 20 µL of the cell suspension in 80 µL of 1X PBS-CMF using a manual counting chamber under a microscope (see Table of Materials). Plate around 40,000 cells/cm2 in each PLL-precoated well in a volume of 1.5 mL of warm AM.
- Seeding of pericytes on the reverted insert filters
- Add 250 µL of collagen type I solution (100 µg/mL) on the reverted insert filters, placed at the periphery of a covered 25 mm high dish (see Table of Materials) using sterile tweezers. Leave the coating for 1 h at RT under sterile conditions.
NOTE: The used dish needs to be high enough to ensure the maintenance of sterility when outside the hood and avoid contact between the solutions on the reverted filter and the cover of the dish. - Remove carefully the collagen type I solution with a glass pipette connected to an aspirating system. Wash twice with 250 µL of RT DMEM HG and then carefully remove all the solution from the insert filters. Leave the coated insert filters at RT under sterile conditions until the seeding of the cells.
NOTE: During the coating procedure, be careful not to touch the filter to avoid membrane damage. Once coated with collagen type I, the insert filters can be stored overnight at RT. - On the day of the triculture setting, wash the pericytes twice with 10 mL of warm 1X PBS-CMF and incubate the cells with 2 mL of warm trypsin. Monitor the action of the trypsin by observing the cells under the microscope. Once the cells start to detach, remove the trypsin and add 5 mL of warm ECM before mechanical dissociation.
- Count the cells by diluting 20 µL of the cell suspension in 80 µL of 1X PBS-CMF using a manual counting chamber under a microscope, and seed 44,500 cells/cm2 on the pre-coated reverted insert filters in a volume of 250 µL. Keep the insert filters in a humidified incubator at 37 °C for 3 h under 5% CO2 and 21% O2.
- Carefully revert the insert filters using sterile tweezers in a 12-well plate containing 1.5 mL of warm ECM/well. The insert filters are now ready to be coated on the other side.
- Add 250 µL of collagen type I solution (100 µg/mL) on the reverted insert filters, placed at the periphery of a covered 25 mm high dish (see Table of Materials) using sterile tweezers. Leave the coating for 1 h at RT under sterile conditions.
- Seeding of endothelial cells on the insert filters
- Coat the upper side of the insert filters with 500 µL of extracellular matrix-based hydrogel (1/48 v/v) (see Table of Materials). After 1 h, in a humidified incubator at 37 °C under 5% CO2 and 21% O2, wash once with 500 µL of RT DMEM HG.
- Wash once with 10 mL of warm 1X PBS-CMF and incubate the cells with 2 mL of warm trypsin. Once the cells start to detach, remove the trypsin and add 5 mL of warm ECM before mechanical dissociation.
- Count the cells by diluting 20 µL of the cell suspension in 80 µL of 1X PBS-CMF using a manual counting chamber under microscope and seed the ECs at a density of 71,500 cells/cm2 on the pre-coated insert filters in a volume of 500 µL of warm ECM.
- Replace AM with 1.5 mL of warm ECM/well and then transfer the seeded insert filters (ECs + pericytes) upon the wells containing the astrocytes.
- Place the triculture cell systems in a humidified incubator at 37 °C under 5% CO2 and 21% O2.
- Maintenance of the triple cell culture for the induction of the BBB properties
NOTE: For the induction of the BBB properties in the ECs, 6 days of triple culture are necessary.- Renew the medium every other day until day 6, taking off carefully the medium from the upper and bottom compartment using a glass pipette connected with an aspiration system.
- Quickly replace with warm ECM in a volume of 500 µL in the upper compartment and 1.5 mL in the bottom compartment, and put back the cells in a humidified incubator at 37 °C under 5% CO2 and 21% O2.
3. BBB phenotype validation
NOTE: After 6 days of triple culture, the time necessary to induce the BBB phenotype in the ECs, the human BBB model is ready for experiments. The physical integrity of the brain-like endothelial cells (BLECs) is visualized by immunofluorescence staining of TJ proteins evaluated using permeability assay to BBB integrity markers. The BBB phenotype validation also includes genes/proteins expression analysis and efflux pumps functionality according to the procedure described in Deligne et al.,23. Pericytes and astrocytes are visualized by respective staining markers according to the procedure described in Deligne et al. 202023.
- Immunofluorescence staining
- Fix the insert filters and astrocytes in ice-cold methanol/acetone (50/50 v/v) for 1 min and wash twice with RT 1X PBS-CMF.
- Carefully separate the filter from the insert by cutting the membrane using a scalpel. Perform immunocytochemistry on the membrane and bottom wells according to Deligne et al.23.
NOTE: For the blocking step, use 250 µL of SEA BLOCK Blocking buffer (see Table of Materials) for 30 min at RT.
- BBB integrity assay
- Assess the physical integrity of the BLECs by a permeability assay using BBB integrity markers with different molecular weights, like Sodium fluorescein (NaF) and 20 kDa Dextran (FD20) (see Table of Materials).
NOTE: The experiment can be performed according to the procedure described in Deligne et al.23.
- Assess the physical integrity of the BLECs by a permeability assay using BBB integrity markers with different molecular weights, like Sodium fluorescein (NaF) and 20 kDa Dextran (FD20) (see Table of Materials).
- Efflux pump functionality
- Assess the functionality of P-glycoprotein (P-gp) and breast cancer resistance protein (BCRP) by measuring the intracellular accumulation of Rhodamine 123 (R123) with and without Elacridar, a P-gp and BCRP inhibitor (see Table of Materials).
NOTE: The experiment can be performed according to the procedure described in Deligne et al.23.
- Assess the functionality of P-glycoprotein (P-gp) and breast cancer resistance protein (BCRP) by measuring the intracellular accumulation of Rhodamine 123 (R123) with and without Elacridar, a P-gp and BCRP inhibitor (see Table of Materials).
- Gene and protein expressions
- Perform gene and protein sample collection on ice after a quick wash with cold Ringer HEPES (RH) (Table 1) of the cells. Before EC sample collection, scrape off the pericytes from the inverted insert filters20.
4. Nanogel transport
NOTE: To estimate the passage of polymeric nanogels (NGs) from the luminal to the abluminal compartment of the triculture BLEC model, 0.1 mg/mL of NG solution was added at day 6 on the luminal compartment for 24 h. Studied NGs were fluorescently tagged N-Isopropylacrylamide (NIPAM)-based hydrogels with an average size of 8-10 nm (see Table of Materials).
- Weigh the nanogel powder and solubilize it in ECM at a concentration of 1 mg/mL. Sonicate the solution for 10 min and filter using a 0.2 µm PTFE filter.
NOTE: Prepare a fresh NG solution the day of the experiment. - Change the medium in the luminal compartment and add 50 µL of NGs solution in the upper compartment for a final concentration of 0.1 mg/mL.
NOTE: Perform a dilution of 1:10 from the original solution. - After 24 h of incubation, collect aliquots from luminal (20 µL) and abluminal (200 µL) compartments and place them in a black 96-well plate.
- Quantify the fluorescence using a fluorescent multiplate reader (see Table of Materials) with a black 96-well plate using the setting of excitation/emission wavelengths at 477/540 nm. Calculate the percentage of crossing referred to the initial working solution added at time = 0 h (t0)6,15.
NOTE: To prepare the 96-well plate for the fluorescence measurement, add 200 µL of solution from the abluminal compartment and 20 µL of solution from the luminal compartment and t0 preparation (add 180 µL of ECM to reach a final volume of 200 µL). Include also a calibration curve and a blank to the reading plate. Instrumental parameters: Detection method – Fluorescence, Optical Position – Top, Read Type – Endpoint, Excitation wavelength – 477 nm, Emission wavelength – 540 nm, Sensitivity – 100, Shake – Double Orbital for 5 s.
Representative Results
Setting of the human triple culture BBB model
The protocol required for the setting of the human BBB in vitro model is described in Figure 1 and includes successive steps whose order must be strictly respected. First, the three cell types are cultivated individually in cell culture dishes (Figure 1A) before being assembled in an insert filter system. The triple culture setting begins with seeding the first cell type, astrocytes, in the pre-coated bottom well. The following day, pericytes and ECs are seeded on the insert filter's pre-coated abluminal and luminal surfaces, respectively. The insert filter is then transferred over the astrocytes. The model is maintained in culture for 6 days, the time necessary to induce the BBB properties in ECs, with a renewal of medium every other day according to the patented co-culture model24. The ECs are then renamed as BLECs (Figure 1B).
Characterization of the human BBB model
The triple cell culture model has been characterized for the presence of a set of BBB-specific properties. First of all, immunocytochemistry data confirmed the expression of conventional markers such as platelet-derived growth factor receptor β (PDGFR-β)25,26 and desmin for pericytes and glial fibrillary acidic protein (GFAP)26 for astrocytes (Figure 2A). Hence, after the 6 days of culture with the pericytes and astrocytes, the monolayer of BLECs, visualized with the adherent junction staining of VE-Cadherin, displays a continuous localization of TJ proteins, Claudin-5 and ZO-1, at the cell borders (Figure 2A). The setting-up of the TJs is correlated with low paracellular permeability coefficients measured using BBB integrity markers of low molecular weight, i.e., NaF (376 Da)16,27 and high molecular weight, i.e., FD20 (20 kDa)27, as shown in Figure 2B. The values measured are comparable with validated BBB in vitro models using the exact source of ECs23,24,28. Altogether, these results highlight the low paracellular permeability of the triple culture BLEC monolayer, which is characteristic of the in vivo BBB. Additionally, R123 intracellular accumulation in BLECs exhibited a significant increase in the presence of the efflux pump inhibitor Elacridar23,24 compared to the control condition with its absence (Figure 2C). This indicates the presence of active efflux pump molecules, namely P-gp and BCRP, in the BLECs.
To further characterize the BLECs, gene expression and protein level of key BBB features were studied (Figure 3). The data obtained with the triple culture model were compared with the validated and patented co-culture model consisting of ECs and pericytes24 used as a control model. The astrocytes represent the third cell type added in the initial co-culture model in the triple culture model. Hence, the gene expression analysis (Figure 3A) of triple culture BLECs, compared with co-culture BLECs, showed the maintenance of expression of key BBB features such as TJ proteins (claudin-5 and zonula occludens-1) and efflux pumps (P-gp and BCRP), and the upregulation of most studied BBB transporters (glucose transporter 1) and receptors (transferrin receptor). Protein quantification data (Figure 3B) were found to be in line with the transcriptional results. Overall, these data support the positive induction of BBB properties in the triple culture BLEC layer similar to the validated co-culture model. Altogether, the triple culture model displays the required physical and metabolic properties for an in vitro microphysiological system to model the BBB.
Applicability to drug delivery strategies – measurement of nanogel transport
To assess the possibility of using the triple culture model to study new brain delivery strategies, the transport of fluorescently-tagged NIPAM-based neutral NGs was evaluated6,15. At time 0, NGs were placed in the luminal compartment at a concentration of 0.1 mg/mL (Figure 4A). After 24 h of incubation, 5.82% of the NGs were found in the abluminal compartment (Figure 4B), proving their ability to cross the BLECs.
The results demonstrate the suitability of the model to measure the permeability of small and larger compounds, as described with the integrity markers, and evaluate the transport of nanomaterials such as polymeric NGs.
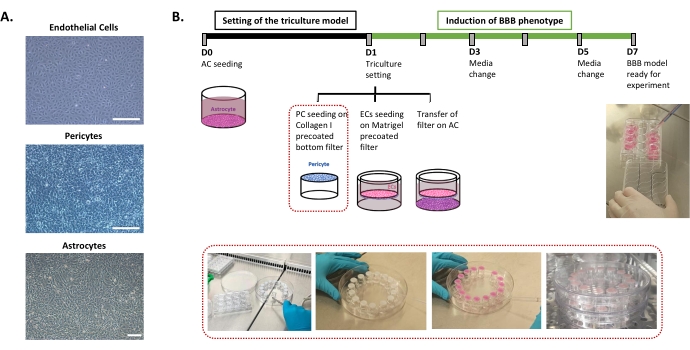
Figure 1: Representation of critical steps for the setting of the triple culture in vitro model of the human BBB. (A) Phase-contrast images of the three cell components of the BBB model: endothelial cells (EC), pericytes (PC), and astrocytes (AC). Scale bar = 250 µm. (B) Schematic and illustrative timeline for the setting of the triple culture human BBB in vitro model. The highlighted box represents the coating procedure for the inverted insert filter. Please click here to view a larger version of this figure.
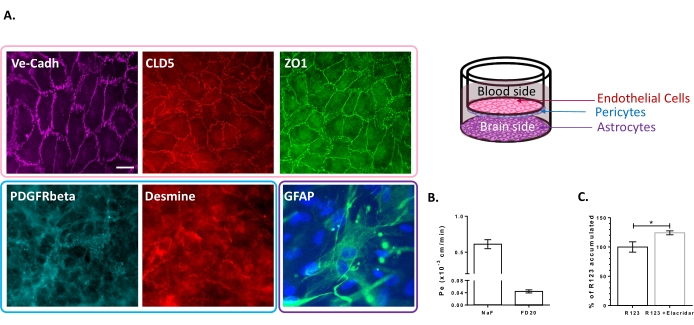
Figure 2: Assessment of the properties of the triple culture BBB model. (A) Representative immunostaining images of the distinctive markers for BLECs (Claudin-5: CLD5, Zona Occludens-1: ZO1 and VE-Cadherin: Ve-Cadh), pericytes (Platelet-Derived Growth Factor Receptor-β: PDGFR-β and desmin), and astrocytes (Glial Fibrillary Acidic Protein: GFAP). Scale bar = 10 µm. (B) Paracellular permeability of BLECs to fluorescent BBB integrity markers, Sodium Fluorescein (NaF, 376 Da, Pe: 0.61 ± 0.062) and FITC-Dextran (FD20, 20 kDa, Pe: 0.04 ± 0.005). N = 3; n = 9. Mean ± SEM. (C) P-gp and BCRP functionality in ECs was assessed by quantifying intracellular R123 with (124.2% ± 3.39%) and without (100% ± 8.79%) Elacridar. N = 4; n = 12. Mean ± SEM. p = 0.017 using an unpaired t-test. Please click here to view a larger version of this figure.
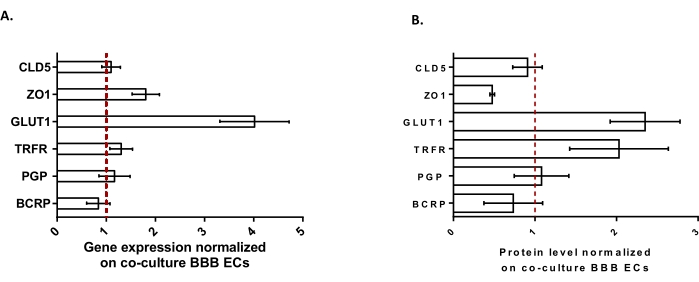
Figure 3: Evaluation of BLEC gene expression and protein level of distinctive markers in the triple culture model compared with the co-culture BBB model. (A) Gene expression of tight junction proteins (Claudin-5: CLD5, and Zona Occludens-1: ZO1), transporters (Glucose Transporter-1: GLUT1, P-glycoprotein: PGP, and Breast Cancer Resistance Protein: BCRP), and large molecule-receptors (Transferrin Receptor: TRFR), normalized by the expression of RPLP0. N = 3; n = 9. (B) Protein level of tight junction proteins (CLD5 and ZO1), transporters (GLUT1, PGP, and BCRP), and large molecule-receptors (TRFR), normalized by the expression of β-actin. N = 3; n = 9. Mean ± SEM. For (A) and (B), values>1 correspond to higher gene expression or protein levels in the triple culture model. The red line corresponds to a value of 1 where the expression level (genes or proteins) of the two models is equivalent. Please click here to view a larger version of this figure.
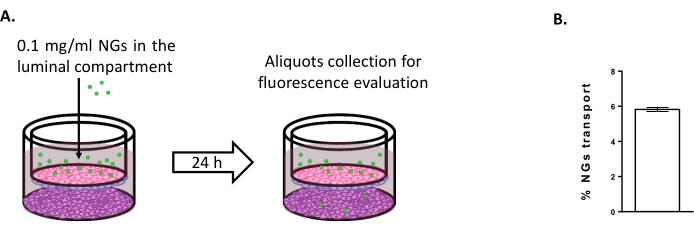
Figure 4: Measuring nanogel transport in the triple culture model. (A) Schematic representation of the nanogel transport assay. (B) Percentage of nanogel transport after 24 h of incubation in the triple culture model (5.82% ± 0.09%). N = 2; n = 6. Mean ± SEM. Please click here to view a larger version of this figure.
| Name of the Buffer | Composition | Note | ||||
| Molecular Weight | ||||||
| Phosphate Buffer Saline, Calcium Magnesium Free | PBS-CMF | NaCl | 8 g/L | 58.4 | Add all the compound to sterile water and wait for complete solubilization. The pH of the obtained solution has to be in the range of 7.3-7.4. Filtrate the solution using a 0.22 µm membrane and store the sterile solution at 4 °C. | |
| KCl | 0.2 g/L | 74.55 | ||||
| KH2PO4 | 0.2 g/L | 136.09 | ||||
| NaHPO4-12 H2O | 2.87 g/L | 358.14 | ||||
| water | ||||||
| Ringer HEPES | RH | NaCl | 8.8 g/L | 58.4 | Add all the compound to sterile water and wait for complete solublization. Adjust the pH to 7.4 (starting solution pH around 6.8). Filtrate the solution using a 0.22 µm membrane and store the sterile solution at 4 °C. | |
| KCl | 0.387 g/L | 74.55 | ||||
| CaCl2 | 0.244 g/L | 110.99 | ||||
| MgCl2 6H2O | 0.0406 g/L | 203.3 | ||||
| NaHCO3 | 0.504 g/L | 84.1 | ||||
| HEPES | 1.19 g/L | 238.3 | ||||
| Glucose | 0.504 g/L | 180.16 | ||||
| water | ||||||
Table 1: Composition of the different buffers used in the protocol.
Discussion
Treatment of brain diseases remains a challenge considering the difficulty of the drugs to hurdle over the BBB to reach their cellular and molecular targets in the brain parenchyma.
Drug development for brain diseases currently exhibits a low success rate since most drugs displaying promising results in preclinical models failed to show any benefit when used in the clinic. Following the "3R rule," which aims at reducing the number of animals used for experimentation, in vitro models of the BBB are developed to study brain pathologies and to predict brain penetration of drugs29. In vitro models of BBB have mainly been developed using animal cells and have become more sophisticated to improve the relevance of the results obtained16. One of the significant advances in the use of human cells, which brings undeniable new insight and more specificity, at the cellular and molecular levels, to study human disease mechanisms16. However, the development of relevant models requires considering the improvement of the BBB in vitro model settings and the knowledge arising, thanks to animal models. Hence, it needs to consider the complexity of the BBB architecture and the importance of the cell-cell communications to study the BBB under physiological and pathological conditions30.
The protocol presented here describes a method to set up a full human BBB in vitro model comprising the three main cell types of the BBB, without limitation of access to brain tissue. As a multiple cell system, the induction and the maintenance of BBB properties, without the artificial use of tightening compounds, but instead induced by cell-cell communications is more physiologically relevant and in line with the in vivo induction of the BBB properties31. Hence, the respect of the chronology of the protocol is prime of importance for the success of the protocol. Moreover, the incubation times during the setting of the triple culture and once the three cell types are assembled represent the main critical steps of the protocol.
The BBB properties in ECs are induced by the co-culture with pericytes, as described for the co-culture model24. Hence, the culture of pericytes at the reverse side of the insert filter is the most critical point and requires strictly following the protocol at the risk of not having enough pericytes for the induction of the BBB properties. First of all, during the coating procedure and also cell seeding, attention has to be put not to have the cover of the Petri dish in contact with the coating and also the medium once the cells are seeded to ensure a good coating of the filter and not to lose cells (steps 2.2.1 and 2.2.4). Moreover, once the pericytes are seeded, it is essential to wait the indicated time for the attachment of the pericytes (step 2.2.4) before reverting the insert filter for the coating and seeding of ECs on the other side (steps 2.2.5 and 2.3). Once seeded, six days are required to induce the BBB properties through cell-cell communications (step 2.4).
The model is validated in terms of restricted permeability (associated with the setting of the tight junctions) since the ECs of the triple culture model display permeability values to BBB integrity markers similar to the validated co-culture model and also measured in validated animal or human models16,27,32. Moreover, the validation of an in vitro BBB model requires, in addition to the restricted permeability, the responsiveness to other cell types of the NVU and the expression of functional receptors and transporters16. In addition, the model is reproducible and produces multiple insert filters and wells to perform numerous analyses (gene and protein expression, fluorescent staining, toxicity tests) on each cell type separately without requiring a cell sorting method.
The model was developed using a 0.4 µm pore size filter to have one cell type on each side of the insert filter. The insert filter system allowed the study of cell-cell communications in physiological conditions by transferring it upon well-containing astrocytes. The presence of astrocytes in the system represents a plus value compared to the initial co-culture in vitro model24. Indeed, considering the importance of astrocytes in the physiology of the BBB, this third cell type allows further understanding of the cell-cell communications within the BBB. Moreover, the triple cell culture system can also be studied in pathological conditions such as stroke, in which the astrocytes play an essential role33,34,35. In addition, the design of BLECs/ pericytes on both sides of the insert filter can easily be placed upon other cell types to mimic pathological conditions such as brain cancer23.
The pore size of the insert filter can bring limitations with some experiments, such as cell transmigration across the BBB. However, the development of the model with a larger pore size requires the adaptation of the protocol to ensure the formation of a physiological monolayer of ECs and not multiple layers, which is not physiologically relevant to mimic the BBB36.
The model's applicability has been demonstrated using NGs transport experiment exhibiting the possibility to do transport experiment using a multicellular system. Nevertheless, one should be aware of the difficulties in having a control compound or molecule for the transport experiment, sharing comparable properties with NGs since each nanostructure exhibits a unique set of properties (molecular weight, charge, shape, physical properties, protein corona formation).
One limitation of the model is the absence of shear stress, which was demonstrated to influence the differentiation of ECs and the expression of TJ proteins37. However, developing a fluidic system mimicking the brain capillary is challenging considering the complexity of adding a fluidic part, requiring a specific device, in a multiple cell system. Moreover, the particular device is usually not commercially available and does not allow many replicates, thus making fluidic systems less adapted for high-throughput use.
In summary, this triple culture system consisting of human cells reproduces in vitro the architecture of the BBB. It allows the generation of many inserts that can be used for extensive screening of compounds.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work is granted by European Union's Horizon 2020 research and innovation program under grant agreement No 764958, as part of the NANOSTEM project, a Marie Skłodowska-Curie Innovative Training Network (ITN) (Fellowship Eleonora Rizzi). This study is granted by the 'Conseil régional du Nord-Pas-de-Calais' (Fellowship to Clémence Deligne), the "Société Française de lutte contre les Cancers et les leucémies de l'Enfant et de l'adolescent"(SFCE), the Association "l'étoile de Martin" and the Association"Cassandra contre la leucémie".
Materials
| Cell Culture | |||
| Astocyte Medium (AM) | ScienCell | 1801 | |
| Astrocyte Growth Supplement | ScienCell | 1852 | Astrocyte Growth Supplement is provided in the AM set. |
| Cell culture dish 100 mm | Corning | 430293 | 100 mm x 20 mm; dish used for the thawing of ECs and PCs before the triculture setting |
| Cell culture dish 150 mm | Corning | 430599 | The height of these dishes (25 mm) allows the seeding of PCs in the reverted insert for the setting of the triculture model. |
| Collagen I | Corning | 354236 | Rat tail |
| Dulbecco's Modified Eagle Medium | Gibco | 31600-083 | Powder |
| Endothelial Cell Growth Supplement | ScienCell | 1051 | Endothelial Cell Growth Supplement is provided in the ECM set. |
| Endothelial Cell Medium (ECM) | ScienCell | 1001 | |
| Fetal Calf Serum | Sigma | F7524 | |
| Gelatin | Sigma | G2500 | 2% gelatin from porcine skin in PBS-CMF |
| Gentamicin | BiochromA6 | A-2712 | |
| Glucose | Sigma | G6152 | Powder |
| Glutamine | Merck | 1002891000 | |
| Human Brain Cortex Astrocytes | ScienCell | 1800-SC | |
| Malassez cell counting chamber | vWR | HECH40453702 | The count was performed manually. |
| Matrigel | Corning | 354230 | Extracellular matrix-based hydrogel |
| Penicillin/Streptomycin | ScienCell | 0503 | Penicillin/Streptomycin solution is provided in the ECM and AM sets. |
| Poly-L-lysine | ScienCell | 0413 | |
| Steritop | Millipore System | SCGPT0SRE | 0.22 µm pore size |
| Transwell insert | Corning | 3401 | 0.4 µm pore polycarbonate filter |
| Trypsin/EDTA neutralization solution | ScienCell | 0113 | |
| Trypsin/EDTA solution | ScienCell | 0103 | |
| Immunocytochemistry | |||
| SEA BLOCK blocking buffer | ThermoScientific | 37527 | |
| Alexa Fluor 568 anti-Mouse secondary antibody | Thermofisher | A11031 | Dilution 1:500 |
| Alexa Fluor 568 anti-Rabbit secondary antibody | Thermofisher | A11036 | Dilution 1:500 |
| Anti-Claudin-5 primary antibody | InVitrogen | 34-1600 | Dilution 1:100 |
| Anti-Desmin primary antibody | Abcam | ab6322 | Dilution 1:200 |
| Anti-Glial Fibrillary Acidic Protein primary antibody | Dako | Z0334 | Dilution 1:500 |
| Anti-Platelet-Derived Growth Factor-β primary antibody | Abcam | ab51090 | Dilution 1:200 |
| Anti-VE-cadherin primary antibody | Abcam | ab207732 | Dilution 1:200 |
| Anti-Zona Occludens-1 primary antibody | InVitrogen | 61-7300 | Dilution 1:200 |
| Normal Goat Serum | Sigma | G6767 | |
| ProLong Diamond Antifade Mountant with DAPI | Invitrogen | P36962 | |
| Gene expression | |||
| NucleoSpin Rna/Protein Macherey Nagel Kit | Macherey-Nagel | 740,933,250 | |
| 96 multiplate well | Biorad | HSP9601 | |
| iSCRIPT | Biorad | 1708841 | |
| Sealer sheet | Biorad | MSB1001 | |
| SsoFast EvaGreen Supermix | Biorad | 172-5201 | |
| Protein expression | |||
| 2x Laemli Sample Buffer | Biorad | 161-0737 | Add 50 µL of bMercaptoetanol to 950 µL of Laemmli Buffer and store at -20°C. Dilute 1:1 with the protein sample for the assay. |
| Anti-Breast Cancer Resistance Protein primary antibody | Abcam | ab207732 | Pre-treatment 15 minutes at RT under agitation; dilution 1:1000, O.N. at 4°C |
| Anti-Claudin-5 primary antibody | Abcam | ab15106 | Pre-treatment 5 minutes at 95°C; dilution 1:1000, O.N. at 4°C |
| Anti-Glucose Transporter 1 primary antibody | Millipore | 07-1401 | Pre-treatment 5 minutes at 95°C; dilution 1:1000, O.N. at 4°C |
| Anti-Mouse secondary antibody | Dako | P0447 | Dilution 1:5000 in TBS-Tween |
| Anti-P-glycoprotein primary antibody | Genetex | GTX23364 | Pre-treatment 15 minutes at RT under agitation; dilution 1:400, 3 hours at RT |
| Anti-Rabbit secondary antibody | Dako | P0448 | Dilution 1:8000 in TBS-Tween |
| Anti-Transferrin Receptor primary antibody | Abcam | ab84036 | Pre-treatment 5 minutes at 95°C; dilution 1:1000, O.N. at 4°C |
| Anti-Zona occludens-1 primary antibody | Abcam | ab216880 | Pre-treatment 5 minutes at 95°C; dilution 1:1000, O.N. at 4°C |
| Criterion TGX Gel | Biorad | 5678083 | |
| ECL Prime Solution | Amersham | RPN2236 | Revelation solution to keep in the dark |
| Phospatase inhibitor cocktail 2 | Sigma | P5726 | |
| Phospatase inhibitor cocktail 3 | Sigma | P0044 | |
| Protease Inhibitor | Sigma | P8340 | |
| Protein Standards | Biorad | 161-0373 | Molecular weight markers |
| RIPA 10x | Millipore | 20-188 | |
| TBS 10x | Biorad | 1706435 | |
| TRIS-Glycine | Biorad | 1610771 | |
| Tween | Biorad | 1706531 | |
| BBB integrity assay | |||
| Sodium Fluorescein | Ampresco | 0681 | λex= 490 nm; λem= 525 nm |
| Elacridar | Sigma | SML0486 | GF120918 |
| FITC-Dextran 20 kDa | Sigma | FD-20S | λex= 490 nm; λem= 525 nm |
| Rhodamine 123 | Sigma | R8004 | λex= 501 nm; λem= 538 nm |
| SynergyTM H1 | BioTek Instruments | Fluorescent multiplate reader | |
| Nanogel Transport | |||
| Syringe | Terumo | SS+01T1 | 1 mL syringe |
| Filter | FisherScientific | 15161499 | 0.2 µm PTFE membrane filter, 15 mm diameter |
| N-Isopropylacrylamide (NIPAM)-based hydrogels | The nanogels (NGs) used in the study are provided by our collaborator in Queen Mary University London, Department of Chemistry. The NGs are covalently tagged with a fluorescent molecule (λex= 477 nm; λem= 540 nm). NGs are freeze dried and shipped as powder, in this state they are stable at room temperature for long period of time. |
References
- Abbott, N. J., Patabendige, A. A., Dolman, D. E., Yusof, S. R., Begley, D. J. Structure and function of the blood-brain barrier. Neurobiology of Disease. 37, 13-25 (2010).
- Profaci, C. P., Munji, R. N., Pulido, R. S., Daneman, R. The blood-brain barrier in health and disease: Important unanswered questions. The Journal of Experimental Medicine. 217 (4), 20190062 (2020).
- Abbott, N. J. Blood-brain barrier structure and function and the challenges for CNS drug delivery. Journal of Inherited Metabolic Disease. 36 (3), 437-449 (2013).
- Kreuter, J. Drug delivery to the central nervous system by polymeric nanoparticles: What do we know. Advanced Drug Delivery Reviews. 71, 2-14 (2014).
- Zhang, W., Mehta, A., Tong, Z., Esser, L. . L.Advanced science. 8 (10), 2003937 (2021).
- Vashist, A., et al. Nanogels as potential drug nanocarriers for CNS drug delivery. Drug Discovery Today. 23 (7), 1436-1443 (2018).
- Lombardo, S. M., Schneider, M., Türeli, A. E., Günday Türeli, N. Key for crossing the BBB with nanoparticles: The rational design. Beilstein Journal of Nanotechnology. 11, 866-883 (2020).
- Bernardo-Castro, S., et al. Therapeutic nanoparticles for the different phases of ischemic stroke. Life. 11 (6), 482 (2021).
- Salinas, Y., Castilla, A. M., Resmini, M. An L-proline based thermoresponsive and pH-switchable nanogel as a drug delivery vehicle. Polymer Chemistry. 9 (17), 2271-2280 (2018).
- Liu, P., Pearce, C. M., Anastasiadi, R. M., Resmini, M., Castilla, A. M. Covalently crosslinked nanogels: An NMR study of the effect of monomer reactivity on composition and structure. Polymers. 11 (2), 353 (2019).
- Preman, N. K., Jain, S., Johnson, R. P. ”Smart” polymer nanogels as pharmaceutical carriers: A versatile platform for programmed delivery and diagnostics. ACS Omega. 6 (8), 5075-5090 (2021).
- Cuggino, J. C., Blanco, E. R. O., Gugliotta, L. M., Alvarez Igarzabal, C. I., Calderón, M. Crossing biological barriers with nanogels to improve drug delivery performance. Journal of Controlled Release Official Journal of the Controlled Release Society. 307, 221-246 (2019).
- Basso, J., et al. Hydrogel-based drug delivery nanosystems for the treatment of brain tumors. Gels. 4 (3), 62 (2018).
- Harilal, S., et al. Revisiting the blood-brain barrier: A hard nut to crack in the transportation of drug molecules. Brain Research Bulletin. 160, 121-140 (2020).
- Papadimitriou, S. A., Robin, M. P., Ceric, D., O’Reilly, R. K., Marino, S., Resmini, M. Fluorescent polymeric nanovehicles for neural stem cell modulation. Nanoscale. 8 (39), 17340-17349 (2016).
- Helms, H. C., et al. In vitro models of the blood-brain barrier: An overview of commonly used brain endothelial cell culture models and guidelines for their use. Journal of Cerebral Blood Flow and Metabolism Official Journal of the International Society of Cerebral Blood Flow and Metabolism. 36 (5), 862-890 (2016).
- Cecchelli, R., et al. Modelling of the blood-brain barrier in drug discovery and development. Nature Reviews Drug Discovery. 6 (8), 650-661 (2007).
- Reichel, A., Begley, D. J., Abbott, N. J. An overview of in vitro techniques for blood-brain barrier studies. Methods in Molecular Medicine. 89, 307-324 (2003).
- Shimizu, F., et al. Peripheral nerve pericytes modify the blood-nerve barrier function and tight junctional molecules through the secretion of various soluble factors. Journal of Cellular Physiology. 226 (1), 255-266 (2011).
- Pedroso, D. C., et al. Improved survival, vascular differentiation and wound healing potential of stem cells co-cultured with endothelial cells. PLoS One. 6 (1), 16114 (2011).
- Kanda, T., Iwasaki, T., Yamawaki, M., Ikeda, K. Isolation and culture of bovine endothelial cells of endoneurial origin. Journal of Neuroscience Research. 49 (6), 769-777 (1997).
- Data sheet on human astrocytes culture. Technical resources from ScienCell Available from: https://www.sciencellonline.com/human-astrocytes.html#product_tabs_technicalresources (2021)
- Deligne, C., et al. Development of a human in vitro blood-brain tumor barrier model of diffuse intrinsic pontine glioma to better understand the chemoresistance. Fluids and Barriers of the CNS. 17 (1), 37 (2020).
- Cecchelli, R., et al. A stable and reproducible human blood-brain barrier model derived from hematopoietic stem cells. PLoS One. 9 (6), 99733 (2014).
- Sweeney, M. D., Zhao, Z., Montagne, A., Nelson, A. R., Zlokovic, B. V. Blood-Brain Barrier: From physiology to disease and back. Physiological Reviews. 99 (1), 21-78 (2019).
- Daneman, R., Prat, A. The Blood-Brain Barrier. Cold Spring Harbor Perspectives in Biology. 7 (1), 020412 (2015).
- Deli, M. A., Ábrahám, C. S., Kataoka, Y., Niwa, M. Permeability studies on in vitro blood-brain barrier models: Physiology, pathology, and pharmacology. Cellular and Molecular Neurobiology. 25 (1), 59-127 (2005).
- Heymans, M., Figueiredo, R., Dehouck, L., Francisco, D., Sano, Y., Shimizu, F., Kanda, T., Bruggmann, R., Engelhardt, B., Winter, P., Gosselet, F., Maxime, C. Contribution of brain pericytes in blood-brain barrier formation and maintenance: A transcriptomic study of co-cultured human endothelial cells derived from hematopoietic stem cells. Fluids Barriers CNS. 17, 48 (2020).
- Russell, W. M. S., Burch, R. L. . The Principles of Humane Experimental Technique. , (1959).
- Neuhaus, W. In Vitro Models of the Blood-Brain Barrier. Handbook of Experimental Pharmacology. 265, 75-110 (2021).
- Hoheisel, D., Nitz, T., Franke, H., Wegener, J., Hakvoort, A., Tilling, T., Galla, H. J. Hydrocortisone reinforces the blood-brain properties in a serum free cell culture system. Biochemical and Biophysical Research Communications. 247 (2), 312-315 (1998).
- Drolez, A., et al. Selection of a relevant in vitro blood-brain barrier model to investigate pro-metastatic features of human breast cancer cell lines. PloS One. 11 (3), 0151155 (2016).
- Neuhaus, W., Gaiser, F., Mahringer, A., Franz, J., Riethmüller, C., Förster, C. The pivotal role of astrocytes in an in vitro stroke model of the blood-brain barrier. Frontiers in Cellular Neuroscience. 8, 352 (2014).
- Mysiorek, C., et al. Peroxisome proliferator-activated receptor-α activation protects brain capillary endothelial cells from oxygen-glucose deprivation-induced hyperpermeability in the blood-brain barrier. Current Neurovascular Research. 6 (3), 181-193 (2009).
- Culot, M., et al. Cerebrovascular protection as a possible mechanism for the protective effects of NXY-059 in preclinical models: An in vitro study. Brain Research. 1294, 144-152 (2009).
- Vandenhaute, E., Drolez, A., Sevin, E., Gosselet, F., Mysiorek, C., Dehouck, M. -. P. Adapting co-culture in vitro models of the blood-brain barrier for use in cancer research: Maintaining an appropriate endothelial monolayer for the assessment of transendothelial migration. Laboratory Investigation. 96 (5), 588-598 (2016).
- Cucullo, L., Hossain, M., Puvenna, V., Marchi, N., Janigro, D. The role of shear stress in Blood-Brain Barrier endothelial physiology. BMC Neuroscience. 12, 40 (2011).

