A Contrast of Three Inoculation Techniques used to Determine the Race of Unknown Fusarium oxysporum f.sp. niveum Isolates
Summary
Managing Fusarium wilt of watermelon requires knowledge of the pathogen races present. Here, we describe the root-dip, infested kernel seeding, and modified tray-dip inoculation methods to demonstrate their efficacy in race-typing of the pathogenic fungus Fusarium oxysporum f. sp niveum (Fon).
Abstract
Fusarium wilt of watermelon (Citrullus lanatus), caused by Fusarium oxysporum f. sp. niveum (Fon), has reemerged as a major production constraint in the southeastern USA, especially in Florida. Deployment of integrated pest management strategies, such as race-specific resistant cultivars, requires information on the diversity and population density of the pathogen in growers’ fields. Despite some progress in developing molecular diagnostic tools to identify pathogen isolates, race determination often requires bioassay approaches.
Race typing was conducted by root-dip inoculation, infested kernel seeding method, and the modified tray-dip method with each of the four watermelon differentials (Black Diamond, Charleston Grey, Calhoun Grey, Plant Introduction 296341-FR). Isolates are assigned a race designation by calculation of disease incidence five weeks after inoculation. If less than 33% of the plants for a particular cultivar were symptomatic, they were categorized as resistant. Those cultivars with incidence greater than 33% were regarded as susceptible. This paper describes three different methods of inoculation to ascertain race, root-dip, infested kernel, and modified tray-dip inoculation, whose applications vary according to the experimental design.
Introduction
The soilborne fungi that make up the Fusarium oxysporum species complex (FOSC) are impactful hemibiotrophic plant pathogens that can cause serious disease and yield loss in a diverse range of crops1. Fusarium wilt of watermelon, caused by F. oxysporum f. sp. niveum (Fon), has been increasing in scope, incidence, and severity across the world in the last several decades2,3. In seedlings, the symptoms of Fusarium wilt often resemble damping-off. In older plants, the foliage becomes gray, chlorotic, and necrotic. Eventually, wilting of the plants progresses to full plant collapse and death4. Direct yield loss occurs due to the symptoms and plant death, while indirect yield loss can occur due to sun damage caused by the elimination of the foliar canopy5. Sexual reproduction and associated reproductive structures have never been observed in F. oxysporum. However, the pathogen produces two types of asexual spores, micro- and macroconidia, as well as larger, long-term survival structures called chlamydospores, which can survive in the soil for many years6.
The FOSC is classified into formae speciales based on observed host ranges, usually limited to one or a few host species1. Although recent research has indicated that this species complex may be a composite of 15 different species, the particular species that infect watermelon are currently unknown7. F. oxysporum f. sp. niveum (Fon) is the name for the groups of strains that exclusively infect Citrullus lanatus or the domesticated watermelon8,9. F. oxysporum strains within most pathogenic formae speciales display certain levels of diversity with regard to their genetic components and virulence toward a host species. For instance, one strain may infect all cultivars of a host, whereas another may only infect the more susceptible cultivars. To account for such variation, these groups are informally classified into races based on evolutionary relationships or common phenotypic characteristics. Within Fon, four races (0, 1, 2, and 3) have been characterized based on their pathogenicity against a set of select watermelon cultivars, with the discovery of race 3 occurring recently10.
Despite this apparent diversity, the morphologies of spores or hyphae are not distinguishable between the races of Fon races, meaning that molecular or phenotypic assays are needed to identify an isolate's unique race11. Molecular research has identified some genetic differences. For example, the role of Secreted in Xylem (SIX) effectors has been studied for years in F. oxysporum, and some of these effectors have been located on the chromosomes exchanged during horizontal gene transfer12. For example, SIX6 is found in Fon races 0 and 1 but not in race 213. SIX effectors have been implicated in the pathogenicity of F. oxysporum f. sp. lycopersici and F. oxysporum f. sp. cubense, which cause Fusarium wilt on tomato and banana, respectively14,15,16,17. The analysis of SIX effector profiles among strains of F. oxysporum f. sp. spiniciae, the Fusarium wilt pathogen on spinach, has enabled classification that accurately reflects genetic and phenotypic diversity18. However, the differences between virulence mechanisms of Fon races are currently not entirely understood, and molecular assays developed upon their use have shown inconsistent and inaccurate results19. Therefore, phenotypic results from infection assays are currently the best way to classify isolates.
F. oxysporum initially infects hosts through the roots before making its way up the xylem20. This makes direct inoculation of the roots of a given host cultivar an effective way to perform race-typing and is the basis of the root-dip and tray-dip inoculation methods21. When not infecting a host, F. oxysporum resides in the soil and can remain dormant for years. Growing susceptible watermelon cultivars in soil from a field of interest is one way to test for the presence of Fon. Expanding this method to include cultivars of different known levels of resistance in soil that is deliberately infested with Fon is also a good way to perform race-typing (Table 1) and is the basis of the infested kernel seeding method. The modified tray-dip method is a variation of the original tray-dip method that allows for a high-throughput race-typing where many plants and field isolates can be investigated rapidly22. Important factors of a quick and successful race-typing bioassay include using cultivars that have documented differences in resistance to the different pathogen races, ensuring that the inoculum is both biologically active and abundant during infection, maintaining an environment that is both conducive for the pathogen and host, and using a consistent rating system for severity or incidence of disease. This paper describes the root-dip23,24, infested kernel seeding25,26, and modified tray-dip22 methods for phenotypic race-typing based on the principles described above.
Protocol
1. Determining race by root-dip method (RDM)
- Preparation of the experimental environment
- Because symptom expression is highly dependent on environmental conditions, maintain plants in a controlled area. Monitor relative humidity, temperature, photoperiod, and light intensity (Figure 1).
- Set the temperature to 26-28 °C, relative humidity to 50-75%, and set a 16 h photoperiod to ensure adequate plant growth and health.
NOTE: To prevent hypoxia, seedling wilting, and/or seed rot, do not overwater or allow standing water around the seedlings. - Use two fluorescent tube lights, each with at least 1850 Lumens of light and a color temperature of 2,800 K per bank of lights to support photosynthetic growth.
- Keep the area clean and use hygienic practices, including the removal of waste soil and plant debris, to prevent pest damage and incidental infection.
- Set the temperature to 26-28 °C, relative humidity to 50-75%, and set a 16 h photoperiod to ensure adequate plant growth and health.
- Planting conditions
- Fill 8 x 16-cell (25 cm width x 50 cm length) starting flats with planting medium and tap down to slightly compress the soil (Figure 2).
- Obtain seeds of the four differential cultivars: Black Diamond/Sugar Baby, Charleston Grey/Allsweet/Dixielee, Calhoun Grey, and PI-296341-FR.
- Sow seeds with their apex (pointed end) pointing upwards to a depth equal to their length. After the seeds are sown, cover the medium containing the buried seeds with 100% Fuller's Earth or other bentonite clay alternative to a depth of 0.3175-0.635 cm.
- Mist the flats to dampen the medium without creating standing or pooling water. Afterwards, keep the media moist by misting for 20 s every 180 min, or watering by hand once per day until seed germination for approximately 5 days. After germination, water once per day and as needed to support the growth of seedlings.
- Media preparation
NOTE: Both media are made firm with extra granulated agar to enable spore harvesting by scraping the surface.- Clarified V8 juice medium (V8)
- Prepare 500 mL of clarified V8 juice medium (V8)27 by adding 100 mL of clarified V8 original 100% vegetable juice with 1% CaCO3 to 400 mL of distilled water.
- Add 7.5 g of granulated agar.
- Mix the ingredients well, autoclave, and allow to cool to 50 °C before pouring into sterile Petri dishes.
- Quarter-strength potato dextrose agar media (qPDA)
- Prepare qPDA medium by adding 4.5 g of granulated agar to 500 mL of distilled water, add 3.8 g of potato dextrose agar.
- Mix the ingredients well, autoclave, and allow to cool to 50 °C before pouring 12-15 mL into sterile Petri dishes.
- Clarified V8 juice medium (V8)
- Because symptom expression is highly dependent on environmental conditions, maintain plants in a controlled area. Monitor relative humidity, temperature, photoperiod, and light intensity (Figure 1).
- Preparation of experimental treatments
- Prepare the inoculum.
- Five days postplanting (dpp), place infiltrated paper disks (1-1.25 cm diameter) containing the preferred F. oxysporum isolate onto one V8 and one qPDA plate and store them in an incubator (~28 °C) for eight days28 (Figure 3A).
- On the eighth day of fungal growth and the day prior to inoculation (see section 1.3.2), transfer the V8 and qPDA plates from the incubator to a biosafety cabinet.
- For each isolate, dispense 6 mL of sterilized deionized water onto each V8 and qPDA culture plate.
- Dislodge conidia by scraping a sterile cell spreader across the medium surface (Figure 3B). Pool the liquid conidial suspension and transfer it to a sterile 50 mL culture tube (Figure 3C).
- Repeat this process until the total liquid conidial suspension volume in the 50 mL culture tube is approximately 12 mL.
- Before proceeding to another isolate, surface-sterilize the work area and cell spreader with alcohol. Sterilize the cell spreader by passing it through a Bunsen burner after dipping it in ≥70% ethanol.
- Once the liquid conidial suspensions have been transferred to culture tubes for all isolates, quantify the spore counts. First, vortex an individual 50 mL culture tube and dispense 10 µL into each chamber of a hemacytometer. Then, calculate the number of spores in the hemacytometer as previously described29.
- Prepare the final inoculum solution by transferring the calculated volume for 106 + 10% to another sterile culture tube and bring the total volume to 30 mL by adding sterile deionized water.
- Store these culture tubes overnight at 8 ± 1 °C.
NOTE: This can be done without loss of conidial viability, as indicated by unpublished data.
- Prepare the inoculum.
- Inoculation
- Prepare the inoculation.
- Prior to the day of inoculation, robustly water thirteen-day-old seedlings to soil carrying capacity.
- Prelabel the stakes with pertinent information, including the isolate to be tested, the watermelon cultivar, and the date of inoculation.
- Place 6 x 12-cell (20 cm width x 40 cm length) styrofoam flats, previously sanitized with 10% bleach and well rinsed, to receive the inoculated plants.
- Preposition all needed materials (see the Table of Materials).
- Inoculate the plant roots.
- Robustly water the plants several hours before beginning the inoculation. At least 2 h after watering, remove the plantlets from the 8 x 16-cell styrofoam flats and rinse their roots to remove any adhering planting material particulates.
- Temporarily store the rinsed plants in clean containers with tap water until use, keeping the cultivars separate from one another (Figure 4A). Separate the plantlets into groups of six individuals and keep the seedling groups wrapped in wet paper towels on a lab tray to prevent desiccation.
- Place 25-30 cm3 of soil in the bottom of each cell of the 6 x 12 array styrofoam tray and use a squirt bottle to wet the soil until it is visibly damp.
- Inoculate the plants and replant them in order according to the cultivar, such that Black Diamond, Charleston Grey, Calhoun Grey, and then PI 296341 01 FR are planted from left to right.
- Beginning with the healthy control, place a group of six undamaged seedlings of the same cultivar into the 50 mL tubes containing the inoculum. In the case of the healthy control, use tap water in lieu of a spore suspension. Inoculate the plants with the positive control (Fon race 3) last.
- Once inside the tube, ensure that the plant roots reach and are exposed to the inoculum (tap water).
- Vortex the tubes with plantlet roots submerged for 30 s (Figure 4B). After vortexing, place a single plantlet per cell in the 6 x 12 styrofoam flats. Place the plants of the same cultivar in the same column in the tray.
- After placement, sanitize gloved hands by holding them in buckets of 0.7% available chlorine solution for 30 s, followed by a tap water rinse for 1 min.
- Afterward, cover the placed plantlets with planting medium and set gently. Using a syringe or pipette, carefully water the plantlets with 20 mL per plantlet while avoiding splashing.
- Before proceeding to the next set of plants, sanitize gloved hands again using chlorine solution and a tap water rinse.
- After all plantlets have been replanted, water them again minimally to prevent inoculum runoff.
- Hold the plantlets overnight in an enclosed, dark environment with an average temperature of 27 °C. The following day, transfer the plants to the greenhouse, keeping the average temperature at 27 °C.
- Prepare the inoculation.
- Maintenance and care of inoculated plants
- To prevent overflow of surplus water, water the flats lightly three times a day for 4-5 days until the plants stabilize.
- Check the trays 2-3 times daily for at least three days to ensure adequate and even coverage of watering.
- Avoid drying due to sunlight or shading by rotating the flats and/or providing supplemental shading/watering as necessary.
- At 3 dpp, fertilize the plants with a 20-20-20 quick-release fertilizer (10 g/3.78 L) at a rate of 3-6 mL per liter of water.
- Fertilize weekly for 3-4 weeks.
- Maintain the same lighting and environmental conditions throughout this step.
2. Determining race by Infested kernel method (IKM)
- Infestation of the kernels
- Prepare the inoculum.
- Either from a stored or newly collected sample, isolate and culture an F. oxysporum f. sp. niveum strain of interest on a plate of qPDA to the point that its growth covers half the plate.
NOTE: This demonstrates that it is active and viable, which is necessary for substantial infestation of the grain in later steps.
- Either from a stored or newly collected sample, isolate and culture an F. oxysporum f. sp. niveum strain of interest on a plate of qPDA to the point that its growth covers half the plate.
- Prepare the kernels.
- On a scale, measure out 200 g of rye (Secale spp.) berries (or Maxie var. wheat (Triticum spp.) kernels) in any sufficiently large container and pour them into one or more 1 L glass Erlenmeyer flasks. Add sterile tap water to the flasks to completely cover the grains up to at least 5 cm (Figure 5A).
- Soak the kernels at room temperature (~24 °C) for 2 h. Drain the water from the flasks; plug the opening with a piece of cotton roll wrapped in cheesecloth; and cover the opening with aluminum foil wrap (Figure 5B).
- Decontaminate the kernels. Once the grain has imbibed water, autoclave it twice in two distinct ways to kill off other unwanted microbes prior to inoculation.
- During the first time, autoclave the grain in the prepared flasks on a gravity cycle (121.2 °C, 1.06 kg/cm2) for 1 h with 5 min drying time. Allow the flasks to cool to room temperature.
- Before autoclaving the second time, transfer the grains to a small mushroom-growing bag with a 0.5 µm filter. Remove the air from the bag and then fold the excess plastic around the bag.
- Place the bag in a plastic, autoclave-safe bin. Cover the bin with aluminum foil wrap (Figure 5C).
NOTE: Do not use a metal bin when autoclaving the grains in the bags, as that may cause the bags to melt. - Autoclave the bin on a gravity cycle for 1 h with 5 min of drying time, the same cycle as before.
NOTE: Only autoclave the bag the second time, not the first, as the filter and port may become compromised if the bags are autoclaved twice. Prevent condensation on the filter by allowing the bags to cool slowly and completely before removing them from the autoclave because the grow bag filter will become compromised if it gets wet.
- Inoculate the kernels.
- Working with the culture plate and the bag in a biosafety cabinet, cut agar discs, 6 mm in diameter, from the zone of active growth on the culture plate with a sterile #4-size cork bore. Unfold the bag. Using a strict sterile technique, place 5 agar discs in the bag. Use a sterile 50 mL graduated cylinder to measure 35 mL of sterile tap water and add it to the bag.
- Roll the opening of the bag somewhat and then spray the outside with 70% ethanol to surface-sterilize.
NOTE: Do not spray the filter as that moisture will also compromise it. - Close the bag by folding the corners towards the center and then over twice above the filter. Secure the bag with bag clamps and clear vinyl tubing provided by the manufacturer.
NOTE: The clamps are reusable. - Remove the bag from the biosafety cabinet.
- Store the pathogen for growth.
- Store the bag upright. Ensure the filter is pulled away from the opposing side of the bag to enable maximum gas exchange (Figure 5D).
- Incubate the materials at room temperature for approximately three weeks. Redistribute the grains regularly to ensure even growth of the pathogen.
NOTE: The bags are not reusable.
- Prepare the inoculum.
- Infection of watermelon seedlings with infested grain
- Source watermelon seeds as previously described.
- Determine the experimental groups.
NOTE: The only strain(s) of the pathogen required are the isolates being tested for race identification. However, negative and positive controls will help make comparisons to plants with no infection or a specific level of infection.- To prepare a negative control, use wheat kernels sterilized according to the previously mentioned method but without inoculation.
- To prepare a positive control, use wheat kernels inoculated with an already classified strain for comparison against the unknown isolate.
- Combine soil and grain.
- Measure 14 grains of infested kernels into a large plastic bag (Figure 5E). Fill plastic pots (15 cm diameter x 10 cm in height) with potting mix to measure the amount of mix needed. Empty the mix into the bag. Create an air cushion in the bag and twist or seal it closed.
- Mix the kernels and soil by inverting the bag several times. For a negative control, perform the same process in a different bag with clean kernels. For a positive control, perform the same process in a different bag with kernels infested with the comparison strain.
- Fill four surface-sterilized pots with the infested soil mixture. For a negative control, pot that mixture before handling any Fon-contaminated soil.
- Sow the watermelon seeds.
- Sow six seeds in each pot. Ensure that each pot only contains seeds from one cultivar. Position the seeds with the apex end of the seed facing up to allow proper growth during emergence (Figure 6A).
- Using a spray bottle, wet the upper 0.3-0.6 cm of soil with water. Place a clear plastic dish (15 cm diameter) under and over each pot to create a humid environment for seed germination (Figure 6B).
- Establish growing conditions.
- Water the pots three times a day with a spray bottle to maintain turgidity without runoff until the seeds have germinated (approximately 5 days).
NOTE: The amount of water used will increase with plant size and pot size. - Once the seeds have germinated, move the top dish to the underside of the pot.
- Water the plants daily as needed for optimal plant growth. Expose the plants to a 16 h photoperiod under similar lighting conditions to those previously described and a temperature of 27 °C (± 1 °C).
- Water the pots three times a day with a spray bottle to maintain turgidity without runoff until the seeds have germinated (approximately 5 days).
3. Determining race by Modified tray-dip method (MTDM)
- Preparation of the materials for inoculations
- Establish planting conditions.
- Fill 48-cell (3.68 cm width x 5.98 cm length x 4.69 cm depth) inserts with steam-pasteurized sand: peat: vermiculite (4:1:1) and tap down to slightly compress the soil. Place the inserts in plastic trays (27.9 cm width x 53.3 cm length x 5.1 cm depth).
- Sow the seeds with their apex (proximal end) pointing upwards to a depth equal to their length.
- Source the seeds as previously described.
- Afterward, keep the media moist by misting for 20 s every 180 min or watering by hand once per day.
- After germination (approximately 5 days), water once per day and as needed to support the growth of the seedlings.
- Prepare the media.
- Prepare qPDA medium by adding 5.625 g of granulated agar to 500 mL of distilled water; add 4.875 g of potato dextrose agar. Mix the ingredients well, autoclave, and cool to 50 °C prior to pouring into sterile Petri dishes. Seal the Petri dishes with parafilm and store the plates in a refrigerator (4 °C) until use.
- To prepare broth medium, weigh and add 24 g of potato dextrose into a 1 L bottle. Bring the mixture to 1000 mL by adding distilled water to the bottle, and place the bottle on a hot plate and stir until dissolved. Dispense 100 mL of the broth into 250 mL Erlenmeyer flasks. Use cheesecloth to seal the flasks and autoclave.
- Prepare the inoculum.
- Seven days before planting, inoculate five qPDA plates with infiltrated paper28 and store them in an incubated area for eight days on a 14 h/10 h dark cycle22.
- On the seventh day, transfer two 1 cm2 agar plugs into each 250 mL Erlenmeyer flask containing 100 mL potato dextrose. Place the Erlenmeyer flasks on a benchtop shaker at 200 rpm for 7 days on a 14 h/10 h light/dark cycle.
- On the day of inoculation (14 days after sowing), harvest the spores by filtering the inoculum through four layers of sterile cheesecloth.
- Determine the microconidial concentration in the flasks using a hemocytometer as previously described. Prepare a 7 L inoculum suspension in a plastic tub (40.6 cm width x 67.3 cm length x 16.8 cm depth) by transferring the correct volume of spore suspension into sterile water for a final spore concentration of 1 × 106 mL−1 (Figure 7A).
- Establish planting conditions.
- Inoculation
- Fourteen days after sowing (at least first true leaf stage), transfer the cell inserts with the seedlings into webbed trays (26.9 cm width x 53.7 cm length x 6.28 cm depth). Gently place the webbed trays with the seedlings into a plastic tub containing the 7 L inoculum suspension. Inoculate each tray one at a time (Figure 7B).
- Allow the seedlings to remain in the inoculum undisturbed for 15 min. After 15 minutes, gently transfer the cell inserts containing the inoculated seedlings into hole-less trays (27.9 cm width x 53.3 cm length x 5.1 cm depth). Repeat this process for each tray.
- Place the hole-less trays on the greenhouse bench and water as needed. Maintain the same lighting and environmental conditions as described for the root-dip bioassay.
4. Disease rating
- Select time intervals for rating.
- For the root-dip and modified tray-dip methods, begin rating one week after the plantlets are inoculated and continue weekly for four more weeks.
NOTE: The combined dataset will include five sets of observations during this period. - While using the kernel method, only begin ratings once the seedlings emerge and continue weekly for a total of six ratings.
- For the root-dip and modified tray-dip methods, begin rating one week after the plantlets are inoculated and continue weekly for four more weeks.
- Observe and calculate the incidence.
- During each rating, take digital images to document the disease progress.
- Report the incidence of wilt and plant death. Calculate the incidence by taking the sum of symptomatic plants compared to the healthy control and the number of dead plants as a proportion of the total number of plants in that cultivar.
- Analyze the results. Compare the patterns of infection between the cultivars and between experiment groups if controls were used.
Representative Results
These experiments help define the relative resistance of commonly grown cultivars (Table 1). This information can then be used to guide management recommendations based on local Fon populations. In other words, if race 0 or 1 is known to be present in a commercial field, then the farmer may be inclined to grow a "resistant" variety such as Calhoun Gray, Sunsugar, or equivalent. The results of the bioassays using all methods show that when the seedlings were infected with a Race 1 isolate, the Black Diamond and Charleston Grey cultivars died or showed serious symptoms, while the Calhoun Grey and PI cultivars showed resistance (Table 2 and Figure 8A).
All methods showed that when the seedlings were infected with a Race 3 isolate, nearly all the plants from all cultivars died or showed serious symptoms (Figure 8B). These results demonstrate how bioassays using both inoculation methods successfully differentiate between races of Fon. The appearance of diseased plants should be the same for all methods. The only difference is in how the cultivars are grouped spatially. For the root-dip and modified dray-dip methods, the cultivars will be organized by columns of the tray, whereas in the kernel method, the cultivars will be grouped in their own pots.

Figure 1: Experimental area for RDM. Due to symptom variability, which is highly dependent on environmental conditions such as relative humidity, temperature, photoperiod, and light intensity, maintaining a regulated experimental area is important. Please click here to view a larger version of this figure.

Figure 2: Preparing the starting flats for RDM. Fill 8 x 16-cell (25 cm width x 50 cm length) starting flats with planting medium and tap down to slightly compress the soil. Please click here to view a larger version of this figure.
Figure 3: Preparation of conidial suspension for RDM. (A) Isolation and culturing. Either from a stored or newly collected sample, isolate and culture a F. oxysporum f. sp. niveum strain of interest on a plate of qPDA to the point that its growth covers half the plate. This demonstrates that it is active and viable, which is necessary for substantial infestation of the grain in later steps. (B) Dislodging conidia. Dislodge conidia by scraping a sterile cell spreader across the medium surface. (C) Suspension deposition. Pool the liquid conidia suspension and transfer it to a sterile 50 mL culture tube. Abbreviation: qPDA = one-quarter strength potato dextrose agar medium. Please click here to view a larger version of this figure.
Figure 4: Organization and vortexing of seedlings for RDM. (A) Separation of cultivars. Temporarily store rinsed plants in clean containers with tap water until use, keeping cultivars separate. (B) Vortexing of seedlings. Vortex the tubes with plantlet roots submerged for 30 s for planting a single plantlet per cell in the 6 x 12 styrofoam flats. Plants of the same cultivar are placed in the same column in the tray. Please click here to view a larger version of this figure.
Figure 5: Preparation and infestation of kernels for IKM. (A) Imbibition of rye berries. On a scale, measure out 200 g of rye (Secale spp.) berries (or Maxie var. wheat (Triticum spp.) kernels) in any sufficiently large container and pour them into one or more 1 L glass Erlenmeyer flasks. Add sterile tap water into the flasks to completely cover the grains up to at least 5 cm. (B) Draining the flasks. Drain the water from the flasks, plug the opening with a piece of cotton roll wrapped in cheesecloth, and cover the opening with aluminum foil wrap. (C) Autoclave setup. Place the bag in a plastic, autoclave-safe bin. Do not use a metal bin when autoclaving the grains in the bags, as that may cause the bags to melt. Cover the bin with aluminum foil wrap. (D) Storage of bags. Store the bag upright. Ensure the filter is pulled away from the opposing side of the bag to enable maximum gas exchange. (E) Measure 14 grains of infested kernels into a large plastic bag. Please click here to view a larger version of this figure.
Figure 6: Sowing and germination of watermelon seeds. (A) Sowing cultivar seeds in pots. Sow six seeds in each pot. Ensure that each pot only contains seeds from one cultivar. Position the seeds with the apex end of the seed facing up to allow proper growth during emergence. (B) Seed germination. Using a spray bottle, wet the upper 0.3-0.6 cm of soil with water. Place a clear plastic dish (15 cm diameter) under and over each pot to create a humid environment for seed germination. Please click here to view a larger version of this figure.
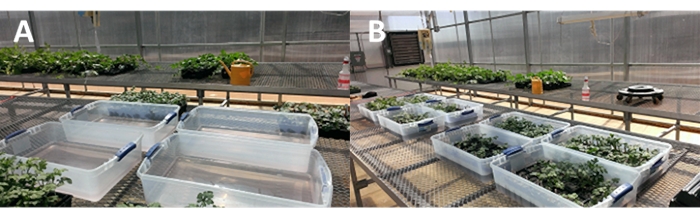
Figure 7: Inoculum preparation and seedling inoculation for MTDM. (A) Preparation of inoculum. Determine the microconidial concentration in the flasks using a hemocytometer as previously described. Prepare a 7 L inoculum suspension in a plastic tub (40.6 cm width × 67.3 cm length × 16.8 cm depth) by transferring the correct volume of spore suspension into sterile water for a final spore concentration of 1 × 106 mL−1. (B) Inoculating the seedlings. Fourteen days after sowing (at least first true leaf stage), transfer the cell inserts with the seedlings into webbed trays (26.9 cm width × 53.7 cm length × 6.28 cm depth). Gently place the webbed trays with the seedlings into a plastic tub containing the 7 L inoculum suspension. Inoculate each tray one at a time. Please click here to view a larger version of this figure.
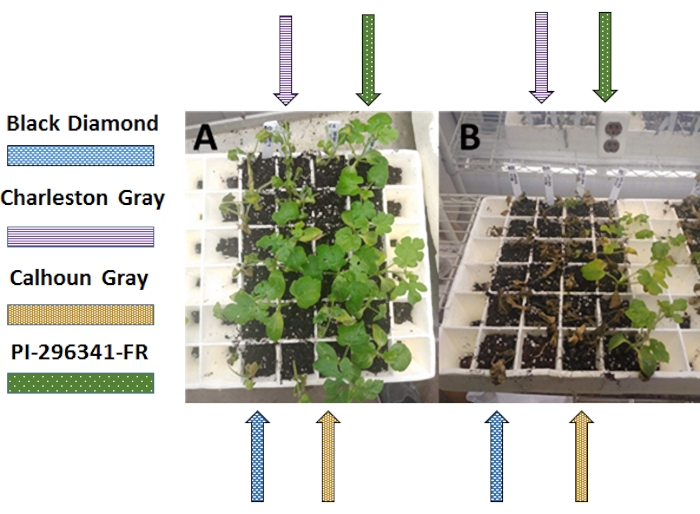
Figure 8: Phenotypic results of race identification methods. (A) Race 1 results. The results of the bioassays using (A) all methods show that when the seedlings were infected with a Race 1 isolate, the Black Diamond and Charleston Grey cultivars died or showed serious symptoms, while the Calhoun Grey and PI cultivars showed resistance. (B) Race 3 results. All methods showed that when the seedlings were infected with a Race 3 isolate, nearly all the plants from all cultivars died or showed serious symptoms. (The appearance of diseased plants should be the same for all methods. Order of planting (left to right) shown by arrows: Black Diamond (blue arrow), Charleston Grey (purple arrow), Calhoun Grey (brown arrow), Plant Introduction 296341-FR (green arrow). Please click here to view a larger version of this figure.
| Cultivar | Race 0 | Race 1 | Race 2 | Race 3 |
| Sugar Baby, Black Diamond | S | S | S | S |
| Charleston Gray, Allsweet, Dixielee | R | S | S | S |
| Calhoun Gray, Sunsugar | R | R | S | S |
| PI-296341-FR | R | R | R | S |
Table 1: Race of Fusarium oxysporum f. sp. Niveum. The race of Fusarium oxysporum f. sp. niveum is determined by susceptible or resistant reactions to a set of watermelon differentials. The cultivars listed in each row are the most used to represent each level of resistance during the evaluation of an isolate's race. This table has been modified from 4. Abbreviations: S = susceptible; R = resistant.
| Isolate | Method | BD | CH. G | Cal G. | PI | Race | ||||
| S | AS | S | AS | S | AS | S | AS | |||
| X | Dip | 6 | 0 | 6 | 0 | 0 | 6 | 0 | 6 | 1 |
| X | Kernel | 6 | 0 | 6 | 0 | 0 | 6 | 0 | 6 | 1 |
| X | MTD | 6 | 0 | 6 | 0 | 0 | 6 | 0 | 6 | 1 |
| Y | Dip | 6 | 0 | 6 | 0 | 6 | 0 | 6 | 0 | 3 |
| Y | Kernel | 6 | 0 | 6 | 0 | 6 | 0 | 6 | 0 | 3 |
| Y | MTD | 6 | 0 | 6 | 0 | 6 | 0 | 6 | 0 | 3 |
| S = Symptomatic; AS = Asymptomatic | ||||||||||
Table 2: Identification of races. Values used in this table reflect incidence or the number of symptomatic plants, compared to the healthy control, and the number of dead plants as a proportion of the total number of plants in that cultivar. The numbers in each cell reflect the incidence reported at the end of the observation period. A cultivar is deemed susceptible when at least 1/3rd or 33% of the plants of that cultivar are symptomatic or dead. The race of the pathogen is then determined based on which cultivars have been deemed susceptible. In other words, how the pathogen performs against cultivars with increasing resistance determines the isolate's race. These results are not from an actual trial and are rather shown to convey how races are identified from the results of these methods. Abbreviations: MTD = modified tray-drip method; BD = Black Diamond; CH. G = Charleston Grey; Cal G. = Calhoun Grey; PI = Plant Introduction 296341-FR; S = symptomatic; AS = asymptomatic.
Discussion
Three methods of race typing have been presented. Each of these methods is best suited to particular questions and experimental conditions. The infested kernel inoculation method (soil infestation) is perhaps simpler and more straightforward, making it especially useful for the assessment of pathogenicity30. Using this method for simple race-typing is highly effective. However, applying the method to determine the resistance of a specific cultivar could be challenging, given that each plant may not face the same degree of infection or exposure, and equally high levels of disease may be needed to test the resistance of the cultivars of interest. This is the case because inoculum produced in this manner is not well quantified, and the proportion of viable propagules, or the number of infectious propagules that reach the root zone, is not well regulated31. Additionally, this method is limited by inconsistencies in the proximity of the planted kernels to the root zone. If too distant, the spores may not germinate, or hyphae may not develop enough to reach the roots.
The root-dip method32,33 is more laborious and time-consuming; however, because the quantity of viable propagules interacting with the plant is more accurately measured, host resistance can be more accurately described, facilitating resistance screening. Moreover, differences in virulence within the same race can be more easily detected. This method has the added benefit that generally, plants become symptomatic earlier and more expressively than in the kernel method. One variant of the root-dip method using chlamydospores in the inoculum suspension instead of conidia may lack this benefit6. Similarly, the modified tray-dip method22 is labor-intensive but allows for high-throughput phenotyping when many isolates and seedlings need to be screened.
Shared factors for the three methods include cultivar selection, growing conditions, and requirements for hygiene. Depending on what is commercially available, certain cultivars can be substituted21,34. Sugar Baby and Black Diamond can both be used to determine race 0 isolates, while Charleston Gray, Allsweet, and Dixielee have been described as resistant to race 0 but susceptible to race 1. Calhoun Gray and Sunsugar are resistant to races 0 and 1 and susceptible to races 2 and 3. Fon disease development is highly dependent on temperature. Care should be taken to ensure that the experimental conditions control for this variable. When choosing a planting medium, general commercial mixes that include peat moss and/or gypsum and allow for good aeration should be satisfactory. Precautions should be taken to prevent cross-contamination of the planting media in both methods, especially from the source bag.
After using one of the described methods, disease must be accurately and consistently assessed. Previous researchers have typically decided on a threshold at which plants are categorized as either susceptible or resistant35,36. For example, if less than 33% of plants of a specific cultivar were symptomatic, then that cultivar would be categorized as resistant with the isolate defined in relation to the susceptible profile of the cultivar. The threshold set is defined by the researcher and the question they wish to address. Variability between raters and by the same rater between plants has been widely reported37,38. Factors such as quality of the seed used, soil quality, inoculum density, storage age of isolates, and rater bias39,40 all contribute to this variability8. Due to this variability in inoculation and PI cultivar responses, multiple experimental replications are needed; ideally, at least three but five replications are recommended, with 6 plants per replication per variety.
While molecular assays have been developed to detect Fon isolates41,42,43, results have not been consistent due to the polyphyletic nature of F. oxysporum and the geographical and genomic variability of the species complex44,45,46. Furthermore, although prior research has established the importance of the Secreted in Xylem (SIX) effectors in full virulence, the exact complement of effectors that define the racial structure of Fon isolates has yet to be determined13. Molecular diagnostics for race are still being developed, for which these phenotypic techniques are critical to assessing their accuracy and utility in Fon race typing19,47.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We would like to acknowledge Dr. Ali and the Plant Molecular Diagnostic Laboratory as well as Dr. Pingsheng Ji at the University of Georgia, whose leadership and support helped establish our Fon program.
Materials
| 100% Fuller’s Earth | Sigma-Aldrich | F200-5KG | |
| 1 L glass Erlenmeyer Flask | PYREX | 4980-1L | |
| 15 mL falcon tubes | Fisher Scientific | 14-959-49B | |
| 50 mL graduated cylinder | Lab Safety Supply | 41121805 | |
| 50 mL Eppendorf Conical Tubes | Fisher Scientific | 05-413-921 | |
| Aluminum foil wrap | Reynolds Wrap | 720 | |
| Bleach | Walmart | 587192290 | |
| Bunsen burner | Fisher Scientific | 03-391-301 | |
| CaCO3 | sigma-Aldrich | 239216 | |
| cell spreaders | Fisher Scientific | 08-100-11 | |
| Cheesecloth | Lions Services, Inc | 8305-01-125-0725 | |
| Clear plastic dishes | Visions Wave | 999RP6CLSS | ~15 cm diameter |
| Clear vinyl tubing for mushroom bag clamps | Shroom Supply | 6" for small bag, 8" for medium bag, 10" for large bag | |
| Cotton Balls | Fisherbrand | 22-456-885 | Sterile |
| Ethanol | Fisher Chemical | A4094 | 100%, then combine with water to make 70% for use |
| Flourescent Tube Lights | MaxLume | Model T5 | 2800 K Color Temperature, 24'' or 48'' long |
| granulated agar | VWR International | 90000-786 | |
| Hand-held Spray Bottle | Ability One | 24122002 | ~0.95 L |
| hemacytometer | Fisher Scientific | 02-671-55A | Two chamber hemacytometer |
| Lab trays | Fisher Scientific | 15-236-2A | |
| Large, sealable plastic bags | Ziploc | 430805 | 38 cm x 38 cm |
| Mister / watering can | Bar5F | B10H22 | |
| Mushroom Bag Clamp | Shroom Supply | 6" for small bag, 8" for medium bag, 10" for large bag | |
| Nitrile Gloves | Fisher Scientific | 19-130-1597D | |
| Organic Rye Berries | Shroom Supply | 0.5 gallon or 25 lb bags | |
| P1000 pipette and tips | Fisher Scientific | 14-388-100 | |
| Petri dishes | Fisherbrand | FB0875713 | Round, 100 mm diameter, 15 mm height |
| Planting media | Jolly Gardener | Pro-Line C/B | |
| Plastic Pitcher | BrandTech | UX0600850 | 1 L or larger |
| Plastic planting pots | Neo/SCI | 01-1177 | ~15 cm diameter and ~10 cm height |
| Plastic, autoclave-safe bin | Thermo Scientific | UX0601022 | 3 L |
| Quarter-strength potato dextrose agar media | Cole-Parmer | UX1420028 | Use powder in combination with recipe for QPDA |
| Scientific Balance Scale, measuring in g | Ohaus | 30208458 | Any precise scale that can hold and measure 200g will work |
| Size #4 cork bore | Cole-Parmer | NC9585352 | |
| Small Mushroom grow bag | Shroom Supply | 0.5 micron filter, also comes in medium and large sizes | |
| Soil trowel | Walmart | 563876946 | |
| Styrofoam flats (6 x 12 cells) | Speedling | Model TR72A | |
| Styrofoam flats (8 x 16 cells) | Speedling | Model TR128A | |
| Syringe (5 or 10 mL) | fisher Scientific | 14-829-19C | |
| Timer | Walmart | TM-01 | |
| V8 Original 100% Vegetable Juice | Walmart | 564638212 | |
| vortex | Fisher Scientific | 02-215-418 | |
| Watermelon Seed – Black Diamond | Willhite Seed Inc | 17 | |
| Watermelon Seed – Calhoun Gray | Holmes Seed Company | 4440 | |
| Watermelon Seed – Charleston Gray | Bonnie Plants | 7.15339E+11 | |
| Watermelon Seed – PI 296341-FR | Contact authors | Contact authors | |
| Wheat Kernels (Maxie var.) (optional) | Alachua County Feed & Seed |
References
- Edel-Hermann, V., Lecomte, C. Current status of Fusarium oxysporum formae speciales and races. Phytopathology. 109 (4), 512-530 (2019).
- Everts, K. L., Himmelstein, J. C. Fusarium wilt of watermelon: Towards sustainable management of a re-emerging plant disease. Crop Protection. 73, 93-99 (2015).
- Martyn, R. Cucurbitaceae 2012. Proceedings of the Xth EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceae. , 136-156 (2012).
- Roberts, P., Dufault, N., Hochmuth, R., Vallad, G., Paret, M. [PP352] Fusarium wilt (Fusarium oxysporum f. sp. niveum) of watermelon. EDIS. 2019 (5), 4 (2019).
- Costa, A. E. S., et al. Resistance to Fusarium wilt in watermelon accessions inoculated by chlamydospores. Scientia Horticulturae. 228, 181-186 (2018).
- Lombard, L., Sandoval-Denis, M., Lamprecht, S. C., Crous, P. Epitypification of Fusarium oxysporum-clearing the taxonomic chaos. Persoonia: Molecular Phylogeny and Evolution of Fungi. 43, 1 (2019).
- Martyn, R. D. Fusarium wilt of watermelon: 120 years of research. Horticultural Reviews. 42 (1), 349-442 (2014).
- Zhou, X., Everts, K. Characterization of a regional population of Fusarium oxysporum f. sp. niveum by race, cross pathogenicity, and vegetative compatibility. Phytopathology. 97 (4), 461-469 (2007).
- Zhou, X., Everts, K., Bruton, B. Race 3, a new and highly virulent race of Fusarium oxysporum f. sp. niveum causing Fusarium wilt in watermelon. Plant Disease. 94 (1), 92-98 (2010).
- Leslie, J. F., Summerell, B. A. . The Fusarium laboratory manual. , (2008).
- Lo Presti, L., et al. Fungal effectors and plant susceptibility. Annual Review of Plant Biology. 66, 513-545 (2015).
- Niu, X., et al. The FonSIX6 gene acts as an avirulence effector in the Fusarium oxysporum f. sp. niveum-watermelon pathosystem. Scientific Reports. 6 (1), 1-7 (2016).
- Lievens, B., Houterman, P. M., Rep, M. Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiology Letters. 300 (2), 201-215 (2009).
- Houterman, P. M., Cornelissen, B. J., Rep, M. Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathogens. 4 (5), 1000061 (2008).
- Houterman, P. M., et al. The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. The Plant Journal. 58 (6), 970-978 (2009).
- Czislowski, E., et al. Investigation of the diversity of effector genes in the banana pathogen, Fusarium oxysporum f. sp. cubense, reveals evidence of horizontal gene transfer. Molecular Plant Pathology. 19 (5), 1155-1171 (2018).
- Batson, A. M., Fokkens, L., Rep, M., du Toit, L. J. Putative effector genes distinguish two pathogenicity groups of Fusarium oxysporum f. sp. spinaciae. Molecular Plant-Microbe Interactions. 34 (2), 141-156 (2021).
- Keinath, A. P., DuBose, V. B., Katawczik, M. M., Wechter, W. P. Identifying races of Fusarium oxysporum f. sp. niveum in South Carolina recovered from watermelon seedlings, plants, and field soil. Plant Disease. 104 (9), 2481-2488 (2020).
- Gordon, T. R. Fusarium oxysporum and the Fusarium wilt syndrome. Annual Review of Phytopathology. 55, 23-39 (2017).
- Martyn, R., Netzer, D. Resistance to races 0, 1, and 2 of Fusarium wilt of watermelon in Citrullus sp. PI-296341-FR. HortScience. 26 (4), 429-432 (1991).
- Meru, G., McGregor, C. Genotyping by sequencing for SNP discovery and genetic mapping of resistance to race 1 of Fusarium oxysporum in watermelon. Scientia Horticulturae. 209, 31-40 (2016).
- Freeman, S., Rodriguez, R. A rapid inoculation technique for assessing pathogenicity of Fusarium oxysporum f. sp. niveum and F. o. melonis on cucurbits. Plant Disease. 77 (12), 1198-1201 (1993).
- Martyn, R. Fusarium oxysporum f. sp. niveum race 2: A highly aggressive race new to the United States. Plant Disease. 71 (3), 233-236 (1987).
- Lai, X., et al. Evaluating inoculation methods to infect sugar beet with Fusarium oxysporum f. Beat and F. secorum. Plant Disease. 104 (5), 1312-1317 (2020).
- Kirk, W., et al. Optimizing fungicide timing for the control of Rhizoctonia crown and root rot of sugar beet using soil temperature and plant growth stages. Plant Disease. 92 (7), 1091-1098 (2008).
- Ferguson, A., Jeffers, S. Detecting multiple species of Phytophthora in container mixes from ornamental crop nurseries. Plant Disease. 83 (12), 1129-1136 (1999).
- Fong, Y., Anuar, S., Lim, H., Tham, F., Sanderson, F. A modified filter paper technique for long-term preservation of some fungal cultures. Mycologist. 14 (3), 127-130 (2000).
- Rice, W. N. The hemocytometer method for detecting fungus spore load carried by wheat. Proceedings of the Association of Official Seed Analysts of North America. 31, 124-127 (1939).
- Kleczewski, N. M., Egel, D. S. A diagnostic guide for Fusarium wilt of watermelon. Plant Health Progress. 12 (1), 27 (2011).
- Dhingra, O. D., Sinclair, J. B. . Basic plant pathology methods. , (2017).
- Latin, R., Snell, S. Comparison of methods for inoculation of muskmelon with Fusarium oxysporum f. sp. melonis. Plant Disease. 70 (4), 297-300 (1986).
- Martyn, R. An iInitial survey of the United States for races of Fursarium oxysporum f. HortScience. 24 (4), 696-698 (1989).
- Zhou, X., Everts, K. Races and inoculum density of Fusarium oxysporum f. sp. niveum in commercial watermelon fields in Maryland and Delaware. Plant Disease. 87 (6), 692-698 (2003).
- Fulton, J. C., et al. Phylogenetic and phenotypic characterization of Fusarium oxysporum f. sp. niveum isolates from Florida-grown watermelon. PLoS One. 16 (3), 0248364 (2021).
- Zhou, X., Everts, K. Quantification of root and stem colonization of watermelon by Fusarium oxysporum f. sp. niveum and its use in evaluating resistance. Phytopathology. 94 (8), 832-841 (2004).
- Nutter, F. W., Esker, P. D., Netto, R. A. C. Disease assessment concepts and the advancements made in improving the accuracy and precision of plant disease data. European Journal of Plant Pathology. 115 (1), 95-103 (2006).
- Nutter, F., Gleason, M., Jenco, J., Christians, N. Assessing the accuracy, intra-rater repeatability, and inter-rater reliability of disease assessment systems. Phytopathology. 83 (8), 806-812 (1993).
- Chiang, K. -. S., Bock, C. H., Lee, I. -. H., El Jarroudi, M., Delfosse, P. Plant disease severity assessment-how rater bias, assessment method, and experimental design affect hypothesis testing and resource use efficiency. Phytopathology. 106 (12), 1451-1464 (2016).
- Nita, M., Ellis, M., Madden, L. Reliability and accuracy of visual estimation of Phomopsis leaf blight of strawberry. Phytopathology. 93 (8), 995-1005 (2003).
- Zhang, Z., Zhang, J., Wang, Y., Zheng, X. Molecular detection of Fusarium oxysporum f. sp. niveum and Mycosphaerella melonis in infected plant tissues and soil. FEMS Microbiology Letters. 249 (1), 39-47 (2005).
- Lin, Y. -. H., et al. Development of the molecular methods for rapid detection and differentiation of Fusarium oxysporum and F. oxysporum f. sp. niveum in Taiwan. New Biotechnology. 27 (4), 409-418 (2010).
- van Dam, P., de Sain, M., Ter Horst, A., vander Gragt, M., Rep, M. Use of comparative genomics-based markers for discrimination of host specificity in Fusarium oxysporum. Applied and Environmental Microbiology. 84 (1), 01868 (2018).
- Baayen, R. P., et al. Gene genealogies and AFLP analyses in the Fusarium oxysporum complex identify monophyletic and nonmonophyletic formae speciales causing wilt and rot disease. Phytopathology. 90 (8), 891-900 (2000).
- O’Donnell, K., Kistler, H. C., Cigelnik, E., Ploetz, R. C. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America. 95 (5), 2044-2049 (1998).
- Laurence, M., Summerell, B., Liew, E. Fusarium oxysporum f. sp. canariensis: evidence for horizontal gene transfer of putative pathogenicity genes. Plant Pathology. 64 (5), 1068-1075 (2015).
- Hudson, O., et al. Marker development for differentiation of Fusarium oxysporum f. sp. Niveum race 3 from races 1 and 2. International Journal of Molecular Sciences. 22 (2), 822 (2021).

