- 00:01Concepts
- 03:13gDNA Isolation and Quality Check
- 03:43Isolation of gDNA and gDNA Quality Check
- 05:52Amplification and Purification of 16S rRNA Gene by PCR
- 07:20Analysis of the DNA Sequences
- 09:02Sequence Assembly and Database Search
16S rRNA测序:基于PCR的技术,用于识别细菌物种
English
소셜에 공유하기
개요
资料来源:埃瓦·布科夫斯卡-法尼班德1号,蒂尔德·安德斯森1号,罗尔夫·卢德1号
1临床科学隆德系,感染医学系,生物医学中心,隆德大学,221 00 隆德,瑞典
地球是数百万种细菌的栖息地,每种细菌物种都有其特殊特征。细菌物种的鉴定在微生物生态学中广泛用于确定环境样本的生物多样性和医学微生物学,以诊断受感染的患者。细菌可以使用传统的微生物学方法进行分类,如显微镜、特定介质的生长、生化和血清学测试以及抗生素敏感性检测。近几十年来,分子微生物学方法彻底改变了细菌鉴定。一个流行的方法是16S核糖体RNA(rRNA)基因测序。该方法不仅比传统方法更快、更准确,而且能够识别在实验室条件下难以生长的菌株。此外,在分子水平上对菌株进行分化,使表性相同的细菌(1-4)之间具有鉴别力。
16S rRNA 与 19 种蛋白质的复合物结合,形成细菌核糖体 (5) 的 30S 亚单位。它由16S rRNA基因编码,由于其在核糖体组装中的基本功能,在所有细菌中都存在并高度保存;然而,它也包含可变区域,可以作为特定物种的指纹。这些特征使16S rRNA基因成为理想的基因片段,可用于细菌的鉴定、比较和遗传分类(6)。
16S rRNA基因测序基于聚合酶链反应(PCR)(7-8),然后是DNA测序(9)。PCR 是一种分子生物学方法,用于通过一系列周期来扩增 DNA 的特定片段,其中包括:
i) 双绞合DNA模板的变性
ii) 与模板互补的引物(短寡核苷酸)的退火
iii) DNA聚合酶的引物延伸,合成新的DNA链
方法的原理图概述如图1所示。
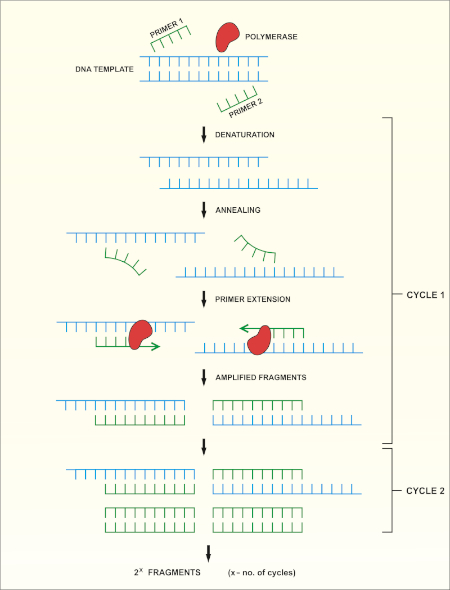
图 1:PCR 反应的原理概述。请点击此处查看此图的较大版本。
有几个因素对成功的PCR反应很重要,其中之一是DNA模板的质量。使用标准协议或商业试剂盒从细菌中分离染色体DNA。应特别注意获得不含污染物的DNA,这些污染物可以抑制PCR反应。
16S rRNA基因的保存区域允许设计通用引物对(一个正向和一个反向),可以结合和放大任何细菌物种的目标区域。目标区域的大小可能有所不同。虽然一些引热剂对可以扩增大多数16S rRNA基因,但其他仅扩增部分。常用引种示例如图1所示,其绑定位点如图2所示。
| 引种名称 | 序列 (5’~3’) | 前进/后退 | 参考 |
| 8F b) | 阿加加特加特格格格格格格格股份公司 | 向前 | -1 |
| 27F | 阿加加特·加·加金茨格格格股份公司 | 向前 | -10 |
| 515F | 格格格格·CMGCCGGGTA | 向前 | -11 |
| 911R | 海合会加特克特科特加特加 | 反向 | -12 |
| 1391R | GACGGGGTGTGTRCA | 反向 | -11 |
| 1492R | GGTTACCTGTTT | 反向 | -11 |
表 1:用于扩增16S rRNA基因a)的标准寡核苷酸的例子。
a)通过计算正向和反向底漆的装订位点之间的距离(见图2),可以估计使用不同引漆组合生成的PCR产品的预期长度,例如PCR的大小使用底漆对 8F-1492R 的产品为 ±1500 bp,对于底漆对 27F-911R =900 bp。
b)也称为 fD1

图 2:16S rRNA序列和引结位点的代表性图。保留区域为灰色,可变区域用对角线填充。为了达到最高分辨率,引基8F和1492R(基于rRNA序列位置的名称)用于放大整个序列,从而允许对基因的几个可变区域进行测序。请点击此处查看此图的较大版本。
PCR 的循环条件(即脱氧核糖核酸变性、引物退火和合成所需的温度和时间)取决于所使用的聚合酶类型和引物的特性。建议遵循制造商针对特定聚合酶的指南。
PCR程序完成后,通过胶质电泳对产品进行分析。成功的 PCR 产生一个预期大小的单个波段。在测序之前,必须对产品进行纯化,以去除PCR反应中存在的残留底漆、脱氧核苷酸、聚合酶和缓冲液。纯化的DNA片段通常被发送到商业测序服务;然而,一些机构在他们自己的核心设施进行DNA测序。
DNA 序列由计算机从 DNA 色谱图自动生成,必须仔细检查质量,因为有时需要手动编辑。按照此步骤,将基因序列与沉积在16S rRNA数据库中的序列进行比较。识别相似区域,并传递最相似的序列。
Procedure
Applications and Summary
Identifying bacterial species is important for different researchers, as well as for those in healthcare. 16S rRNA sequencing was initially used by researchers to determine phylogenetic relationships between bacteria. In time, it has been implemented in metagenomic studies to determine biodiversity of environmental samples and in clinical laboratories as a method to identify potential pathogens. It enables a quick and accurate identification of bacteria present in clinical samples, facilitating earlier diagnosis and faster treatment of patients.
References
- Weisburg, W.G., Barns, S.M., Pelletier, D.A. and Lane D.J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 173 (2): 697-703. (1991)
- Drancourt, M., Bollet, C., Carlioz, A., Martelin, R., Gayral, J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 38 (10):3623-3630. (2000)
- Woo, P.C., Lau, S.K., Teng, J.L., Tse, H., Yuen, K.Y. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect. 14 (10):908-934. (2008)
- Tang, Y.W., Ellis, N.M., Hopkins, M.K., Smith, D.H., Dodge, D.E., Persing, D.H. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J Clin Microbiol. 36 (12):3674-3679. (1998)
- Tsiboli, P., Herfurth, E., Choli, T. Purification and characterization of the 30S ribosomal proteins from the bacterium Thermus thermophilus. Eur J Biochem. 226 (1):169-177. (1994)
- Woese, C.R. Bacterial evolution. Microbiol Rev. 51 (2):221-271. (1987)
- Bartlett, J.M., Stirling, D. A short history of the polymerase chain reaction. Methods Mol Biol. 226:3-6. (2003)
- Wilson, K.H., Blitchington, R.B., Greene, R.C. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 28 (9):1942-1946. (1990)
- Shendure, J., Balasubramanian, S., Church, G.M., Gilbert, W., Rogers, J., Schloss, J.A., Waterston, R.H. (2017) DNA sequencing at 40: past, present and future. Nature. 550:345-353.
- Lane, D.J. 16S/23S rRNA sequencing. (1991) In Nucleic acid techniques in bacterial systematics. (Goodfellow, M. and Stackebrandt, E., eds.) p.115-175. Wiley and Sons, Chichester, United Kingdom.
- Turner, S., Pryer, K.M., Miao, V.P., Palmer, J.D. (1999) Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol. 46:327-338.
- Fredricks, D.N., Relman, D.A. (1998) Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J Clin Microbiol. 36:2810-2816.
- Wilson, K. Preparation of genomic DNA from bacteria. (2001) Curr Protoc Mol Biol. Chapter 2:Unit 2.4.
- Wright, M. H., Adelskov, J., Greene, A.C. (2017) Bacterial DNA extraction using individual enzymes and phenol/chloroform separation. J Microbiol Biol Educ. 18:18.2.48.
- Huang, X., Madan, A. (1999). CAP3: A DNA sequence assembly program. Genome Res. 9:868-877.
내레이션 대본
Earth is home to millions of bacterial species, each with unique characteristics. Identifying these species is critical in evaluating environmental samples. Doctors also need to distinguish different bacterial species to diagnose infected patients.
To identify bacteria, a variety of techniques can be employed, including microscopic observation of morphology or growth on a specific media to observe colony morphology. Genetic analysis, another technique for identifying bacteria has grown in popularity in recent years, due in part to 16S ribosomal RNA gene sequencing.
The bacterial ribosome is a protein RNA complex consisting of two subunits. The 30S subunit, the smaller of these two subunits, contains 16S rRNA, which is encoded by the 16S rRNA gene contained within the genomic DNA. Specific regions of 16S rRNA are highly conserved, due to their essential function in ribosome assembly. While other regions, less critical to function, may vary among bacterial species. The variable regions in 16S rRNA, can serve as unique molecular fingerprints for bacterial species, allowing us to distinguish phenotypically identical strains.
After obtaining a quality sample of gDNA, PCR of the 16S rRNA encoding gene can begin. PCR is a commonly used molecular biology method, consisting of cycles of denaturation of the double-stranded DNA template, annealing of universal primer pairs, which amplify highly conserved regions of the gene, and the extension of primers by DNA polymerase. While some primers amplify most of the 16S rRNA encoding gene, others only amplify fragments of it. After PCR, the products can be analyzed via agarose gel electrophoresis. If amplification was successful, the gel should contain a single band of an expected size, depending upon the primer pair used, up to 1500bp, the approximate length of the 16S rRNA gene.
After purification and sequencing, the obtained sequences can then be entered into the BLAST database, where they can be compared with reference 16S rRNA sequences. As this database returns matches based on the highest similarity, this allows confirmation of the identity of the bacteria of interest. In this video, you will observe 16S rRNA gene sequencing, including PCR, DNA sequence analysis and editing, sequence assembly and database searching.
When handling microorganisms, it is essential to follow good microbiological practice, including using aseptic technique and wearing appropriate personal protective equipment. After performing an appropriate risk assessment for the microorganism or environmental sample of interest, obtain a test culture. In this example, a pure culture of Bacillus subtilis is used.
To begin, grow your microorganism on a suitable medium in the appropriate conditions. In this example, Bacillus subtilis 168 is grown in LB broth overnight in a shaking incubator set to 200 rpm at 37 degrees Celsius. Next, use a commercially available kit to isolate genomic DNA or gDNA from 1.5 milliliters of the B. subtilis overnight culture.
To check the quality of the isolated DNA, first mix five microliters of the isolated gDNA with one microliter of DNA gel loading dye. Then, load the sample onto a 0.8% agarose gel, containing DNA staining reagent, such as SYBR safe or EtBr. After this, load a one kilobase molecular mass standard onto the gel, and run the electrophoresis until the front dye is approximately 0.5 centimeters from the bottom of the gel. Once the gel electrophoresis is complete, visualize the gel on a blue light transilluminator. The gDNA should appear as a thick band, above 10 kilobase in size and have minimal smearing.
After this, to create serial dilutions of the gDNA, label three microcentrifuge tubes as 10X, 100X, and 1000X. Then, use a pipette to dispense 90 microliters of sterile distilled water into each of the tubes. Next, add 10 microliters of the gDNA solution to the 10X tube. Pipette the whole volume up and down to ensure the solution is mixed thoroughly. Then, remove 10 microliters of the solution from the 10X tube and transfer this to the 100X tube. Mix the solution as previously described. Finally, transfer 10 microliters of the solution in the 100X tube, to the 1000X tube.
To begin the PCR protocol, thaw the necessary reagents on ice. Then, prepare the PCR master mix. Since the DNA polymerase is active at room temperature, the reaction set up must occur on ice. Aliquot 49 microliters of the master mix into each of the PCR tubes. Then, add one microliter of template to each of the experimental tubes and one microliter of sterile water to the negative control tube, pipetting up and down to mix. After this, set the PCR machine according to the program described in the table. Place the tubes into the thermocycler and start the program.
Once the program is complete, examine the quality of your product via agarose gel electrophoresis, as previously demonstrated. A successful reaction using the described protocol should yield a single band of approximately 1.5 kilobase. In this example, the sample containing 100X diluted gDNA yielded the highest quality product. Next, purify the best PCR product, in this case, the 100X gDNA, with a commercially available kit. Now the PCR product can be sent for sequencing.
In this example, the PCR product is sequenced using forward and reverse primers. Thus, two data sets, each containing a DNA sequence and a DNA chromatogram, are generated: one for the forward primer and the other for the reverse primer. First, examine the chromatograms generated from each primer. An ideal chromatogram should have evenly spaced peaks with little to no background signals.
If the chromatograms display double peaks, multiple DNA templates may have been present in the PCR products and the sequence should be discarded. If the chromatograms contained peaks of different colors in the same location, the sequencing software likely miscalled nucleotides. This error can be manually identified and corrected in the text file. The presence of broad peaks in the chromatogram indicates a loss of resolution, which causes miscounting of the nucleotides in the associated regions. This error is difficult to correct and mismatches in any of the subsequent steps should be treated as unreliable. Poor chromatogram reading quality, indicated by the presence of multiple peaks, usually occurs at the five prime and three prime ends of the sequence. Some sequencing programs remove these low quality sections automatically. If your sequence was not truncated automatically, identify the low quality fragments and remove their respective bases from the text file.
Use a DNA assembly program to assemble the two primer sequences into one continuous sequence. Remember, sequences obtained using forward and reverse primers should partially overlap. In the DNA assembly program, insert the two sequences in FASTA format into the appropriate box. Then, click the submit button and wait for the program to return the results.
To view the assembled sequence, click on Contigs in the results tab. Then, to view the details of the alignment, select assembly details. Navigate to the website for the basic local alignment search tool, or BLAST, and select the nucleotide BLAST tool to compare your sequence to the database. Enter your sequence into the query sequence text box and select the appropriate database in the scroll down menu. Finally, click the BLAST button on the bottom of the page, and wait for the tool to return the most similar sequences from the database.
In this example, the top hit is B. subtilis strain 168, showing 100% identity with the sequence in the BLAST database. If the top hit does not show 100% identity to your expected species or strain, click on the sequence which most closely matches your query to see the details of the alignment. Aligned nucleotides will be joined by short vertical lines and mismatched nucleotides will have gaps between them. Focusing on the identified mismatched regions, revise the sequence and repeat the BLAST search if desired.
