Establishing Mixed Neuronal and Glial Cell Cultures from Embryonic Mouse Brains to Study Infection and Innate Immunity
Summary
This protocol presents a unique way of generating central nervous system cell cultures from embryonic day 17 mouse brains for neuro(immuno)logy research. This model can be analyzed using various experimental techniques, including RT-qPCR, microscopy, ELISA, and flow cytometry.
Abstract
Models of the central nervous system (CNS) must recapitulate the complex network of interconnected cells found in vivo. The CNS consists primarily of neurons, astrocytes, oligodendrocytes, and microglia. Due to increasing efforts to replace and reduce animal use, a variety of in vitro cell culture systems have been developed to explore innate cell properties, which allow the development of therapeutics for CNS infections and pathologies. Whilst certain research questions can be addressed by human-based cell culture systems, such as (induced) pluripotent stem cells, working with human cells has its own limitations with regard to availability, costs, and ethics. Here, we describe a unique protocol for isolating and culturing cells from embryonic mouse brains. The resulting mixed neural cell cultures mimic several cell populations and interactions found in the brain in vivo. Compared to current equivalent methods, this protocol more closely mimics the characteristics of the brain and also garners more cells, thus allowing for more experimental conditions to be investigated from one pregnant mouse. Further, the protocol is relatively easy and highly reproducible. These cultures have been optimized for use at various scales, including 96-well based high throughput screens, 24-well microscopy analysis, and 6-well cultures for flow cytometry and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis. This culture method is a powerful tool to investigate infection and immunity within the context of some of the complexity of the CNS with the convenience of in vitro methods.
Introduction
Improving our understanding of the central nervous system (CNS) is critical to improve therapeutic options for many neuroinflammatory and neurodegenerative diseases. The CNS, a complex network of interconnected cells within the brain, spinal cord, and optic nerves, comprises neurons, oligodendrocytes, astrocytes, and their innate immune cells, the microglia1. An in vitro approach can often drastically reduce the number of mice required to perform meaningful research; however, the complex nature of the CNS makes it impossible to recapitulate the in vivo situation using cell lines. Mixed neural cell cultures provide an extremely valuable research tool to investigate neuro(immuno)logy questions in a relevant model, in line with the Replacement, Reduction and Refinement (3Rs) principles2,3.
Thomson et al. described a cell culture method using prenatal spinal cord cells that differentiate into all the aforementioned main CNS cell types4. This system also has synapse formation, myelinated axons, and nodes of Ranvier. The main limitation of this culturing method is that, being spinal cord, it does not usefully model the brain, and the cell yields from embryonic day 13 (E13) spinal cords are constricting. Thus, this limits the number of experimental conditions that can be investigated. Therefore, this study aimed to develop a new cell culture system that recapitulates the characteristics of the brain with increased cell yield to reduce the requirements for animals.
Using Thomson et al. as a starting point, we developed a cell culture model derived purely from prenatal mouse brains. These cultures have the same cell populations, interconnectivity, and treatment options as the spinal cord cultures, except there is less myelination by comparison. However, having a CNS in vitro model with an approximately threefold higher cell yield is more efficient, requiring fewer mice and less time processing embryos. We optimized this unique culture system for multiple downstream applications and scales, including using glass coverslips for microscopy analysis and various sizes of plastic well plates, including 96-well plates for high throughput research.
Protocol
All animal experiments complied with local laws and guidelines for animal use, and were approved by the local Ethical Review Committee at the University of Glasgow. Animals were housed in specific pathogen-free conditions in accordance with the UK Animals Scientific Procedures Act 1986, under the auspices of a UK Home Office Project License. For this study, in-house bred adult C57BL/6J mice were used. The use of young females (8-12 weeks) is recommended due to the higher success rate of pregnancy; males can be reused for multiple rounds of breeding. Figure 1 represents a schematic overview of the described method to generate mixed neuronal and glial cultures.
1. Preparation of the tissue culture consumables
- Prepare the plates and/or dishes containing microscopy coverslips inside a Class 2 Safety cabinet. Sterilize all reagents or autoclave to ensure sterility during the culture period.
- Add the appropriate volume of BA-PLL (13.2 µg/mL poly-L-lysinehydrobromide [PLL] in boric acid buffer [BA] [50 mM boric acid, 12.5 mM sodium tetraborate, pH 8.5]; see Table of Materials) to each well (1,000 µL/well in a 6-well plate, 100 µL/well in a 96-well plate; volumes are summarized in Table 1). For microscopy coverslips, add 20 mL of BA-PLL to a 9 cm diameter tissue culture dish containing 200 sterile coverslips, and swirl to distribute evenly.
- Incubate at 37 °C for 1-2 h.
- Remove the BA-PLL solution from each well or dish containing microscopy coverslips, and wash by adding 20 mL of sterile water, swirling the coverslips, and then removing the water. Repeat this wash step three times. For a dish containing coverslips, leave sterile water in the tissue culture dish on the final wash to easily remove the coverslips.
- Remove as much liquid as possible with a sterile pipette and allow to dry for at least 2 h to overnight.
- Store the coated plates at 4 °C for up to 2 months.
NOTE: The prepared boric acid with poly-l-lysine (BA-PLL) solution can be reused up to three time, adding new PLL each time. Store the BA-PLL at 4 °C. Dishes are treated with BA-PLL, as the PLL allows the cells to stick down and grow. Without this treatment, the cells will lift after approximately 1 week of culture and no longer be able to differentiate.
2. Dissection of E17 embryonal brains
- Co-house one or multiple female mice with a male mouse. Check the females daily for a mucus plug, indicating mating has taken place.
NOTE: Any "plugged" female mice need to be separated from the male to ensure the correct start date of gestation. Mice can be weighed to confirm pregnancy or monitored visually. - Cull the pregnant mouse at E17 using appropriate methods in compliance with local animal welfare guidance and laws, for example, by raising carbon dioxide (CO2) concentration, a lethal anesthetic overdose, or dislocation of the neck.
NOTE: The chosen method must not disrupt the embryos. For this study, exposure to a rising concentration of carbon dioxide gas followed by confirmation of death by severing the femoral artery was used to cull pregnant dams. - Place the culled, pregnant mouse on its back on a dissection board; while pinning it down is not required, it might make it easier for inexperienced researchers. Pinch the midline of the abdomen using forceps. Using sharp scissors, cut open the abdomen through the skin and the peritoneum over the midline from the genitalia to the ribcage, being careful not to puncture the uterus.
- The mouse uterus has two horns, each typically containing one to five embryos. Remove the uterus containing the embryos from the mother and immediately place it on ice.
- Cut through the yolk sack on the side of the placentas, being careful not to damage the embryos, and remove the embryos from their yolk sack.
- Immediately decapitate the embryos. Add the decapitated heads into a dish with Hanks' balanced salt solution (HBSS) without calcium (Ca2+) and magnesium (Mg2+) (HBSS-/-) on ice.
NOTE: If genotyping is required, one can remove the tail at this stage for genetic analysis. When multiple genotypes are expected, the heads of each embryo should be kept separately for culturing. - Using angled forceps, position the head on its side facing left.
- Pierce the eye with one edge of the forceps, firmly holding the chin with the other.
- Starting at the nape, gently tear the skin of the scalp along the midline toward the tip of the snout.
- Entering through the spinal cord, noticeable as a white oval, use the angled forceps to crack open the skull along the midline, exposing the brain.
- Gently peel the skull away on the side facing upward, exposing the brain.
- Lift the brain out of the skull, disposing of the skull once the brain has been completely removed.
- Using the forceps, remove the meninges, which are noticeable as a thin membrane with dense blood vessels.
- Place the brains into a bijou (see Table of Materials) containing 2 mL of HBSS-/- on ice.
- Repeat steps 2.6 to 2.13 with the remaining brains, adding up to four brains per bijou.
- Add 250 µL of 10x trypsin to the bijou and triturate the brains by shaking the bijou. Incubate for 15 min at 37 °C.
NOTE: All steps from this point forward should be performed in a sterile tissue culture hood. - Thaw 2 mL of soybean trypsin (SD) inhibitor (Leibovitz L-15, 0.52 mg/mL trypsin inhibitor from soybean, 40 µg/mL DNase I, 3 mg/mL bovine serum albumin [BSA] fraction V; see Table of Materials) from -20 °C by placing it at 37 °C.
- Add 2 mL of SD inhibitor to each bijou containing brains (per bijou containing up to four brains), shaking the bijou again to disperse it evenly.
NOTE: SD inhibitor decreases the trypsin activity to prevent unnecessary digestion of the samples and preserve cell viability. - Without centrifugation, remove 2 mL of the supernatant from each bijou and transfer into a 15 mL centrifuge tube, being careful not to transfer cell clumps.
- Triturate the remaining cells in the bijou with a 19 G needle attached to a 5 mL syringe by aspirating the suspension twice. This will create a thick mucus-like mixture.
- Repeat twice more using a 21 G needle. If there are clumps remaining, triturate once more with the 21 G needle.
- Transfer the cells from the bijou to the same 15 mL centrifuge tube (from step 2.18) using a 23 G needle.
- Centrifuge at 200 x g at room temperature (RT) for 5 min.
- Remove all the supernatant using a 5 mL serological pipette and transfer it to another 15 mL centrifuge tube, being careful not to disturb the loose pellet at the bottom containing the required cells.
- Centrifuge the supernatant again at 200 x g at RT for 5 min.
NOTE: This step is not essential, but if one requires many cells or has few embryos, one could perform this step to recover as many cells as possible from the supernatant. - Using 10 mL of plating media (PM) (49% Dulbecco's modified eagle medium [DMEM], 1% penicillin/streptomycin [Pen/Strep], 25% horse serum, 25% HBSS with Ca2+ and Mg2+ [HBSS+/+]; see Table of Materials), combine and resuspend the two pellets together to create a whole cell suspension.
- Count the cells using trypan blue and either a hemocytometer or digital cell counter, and dilute the cell suspension with PM to a concentration of 1.8 x 106 cells/mL.
3. Plating the cells
- Add the required volume of the cell suspension to the required format as detailed in Table 1: 1,000 µL per well in 6-well format, 50 µL per well in 96-well format, or 100 µL per coverslip.
- Incubate for 2-4 h at 37 °C with 5%-7% CO2. Check the cells have adhered using an inverted microscope.
- Remove the media and top up with new differentiation media (DM+: DM- including 10 µg/mL insulin. DM-: DMEM, 1% Pen/Strep, 50 nM hydrocortisone, 10 ng/mL biotin, 2.5 mL 100x N1 media supplement; see Table of Materials). Volumes are detailed in Table 1. Press down any floating coverslips using a sterile pipette tip.
4. Maintaining the cultures
NOTE: These cultures require feeding thrice weekly to support optimal growth and differentiation. The cultures will reach optimum health and maturity for experiments on DIV (days in vitro) 21. Cells can be kept in culture for up to 28 days, after which the cultures quickly degenerate.
- Three times per week until DIV12, replace part of the supernatant with fresh DM+ by removing 500 µL per well in 6-well format, 50 µL per well in 96-well format, or 500 µL per dish containing three coverslips, and adding 600 µL per well in 6-well format, 60 µL per well in 96-well format, or 600 µL per coverslip dish (Table 1).
- Three times per week from DIV13 onwards, replace part of the supernatant with fresh DM- by removing 500 µL per well in 6-well format, 50 µL per well in 96-well format, or 500 µL per dish containing three coverslips, and adding 600 µL per well in 6-well format, 60 µL per well in 96-well format, or 600 µL per coverslip dish (Table 1).
Representative Results
Microscopy
Cultures grown on glass coverslips are ideal to analyze by microscopy. To visualize the development of the cultures, the coverslips were fixed in 4% paraformaldehyde (PFA) at multiple timepoints from DIV0 (once cells were attached) until DIV28. The cultures were stained for immunofluorescence imaging as previously described5 using three different staining combinations: NG2 (immature oligodendrocytes) and nestin (neuronal stem/progenitor cells) as developmental markers, SMI31 (axons), MBP (myelin), and NeuN (neuron cell body) as neuronal markers, or CNP (oligodendrocytes), GFAP (astrocytes), and Iba1 (microglia) as glial markers (Figure 2).
The various cell types were quantified using CellProfiler pipelines (based on ′,6-diamidino-2-phenylindole-positive (https://github.com/muecs/cp/tree/v1.1). Each individual data point was generated from an average of 10 images taken from three coverslips per timepoint. For microglia, neurons, and oligodendrocytes, the percentage of cell type per 4′,6-diamidino-2-phenylindole-positive (DAPI+) cell was calculated. The percentage field of view was used instead of the number of cells for quantifying the number of astrocytes. This was due to difficulties differentiating between the individual cells as they frequently overlapped (Figure 2M,N). The cultures reached peak maturity and cell density at DIV21, after which the cultures started to degrade (Figure 2N).
Importantly, these cultures can easily be treated with drugs, such as potential therapeutics, or used to trace in vitro infections. In this example, cultures on coverslips were transferred to a 24-well plate and infected with the highly neurotropic virus Semliki Forest virus (SFV) (strain SFV6)6, which expresses zsGreen in infected cells. A multiplicity of infection (MOI-the number of virus particles added per cell) of 0.05, as titrated on baby hamster kidney (BHK) cells, was used to ensure low-level infection. After 0-72 h post-infection (hpi), cultures were fixed in 4% PFA and stained for analysis by immunofluorescent imaging. Figure 3 illustrates that, in line with in vivo infection, SFV mainly infects oligodendrocytes and neurons6.
qRT-PCR
In addition to microscopy, these CNS cultures can be used for analysis by molecular methods, such as RT-qPCR of mRNA responses to treatment. To further investigate the innate antiviral response, 6-well plate cultures were treated with a range of doses of the potent antiviral cytokine interferon beta (IFN-β) for 24 h. Cultures were lysed with guanidium-thiocyanate/phenol, and the RNA was isolated, converted to cDNA, and analyzed by qPCR as previously described7. Using this method, differential expression of many genes can be measured. Here, the upregulation of Ccl5 was quantified against the housekeeping gene 18s. CCL5 is a chemotactic cytokine (chemokine) involved in the inflammatory response. IFN-β treatment resulted in an upregulation of Ccl5 mRNA in the cultures (Figure 4A).
Enzyme-linked immunosorbent assay (ELISA)
The 96-well format of cultures is an excellent tool for high throughput screenings of treatments. To investigate whether the upregulation of Ccl5 mRNA results in the expression of CCL5 protein, CCL5 was measured in the supernatant of the cultures by ELISA, according to the manufacturer's instructions (see Table of Materials). In this example, 96-well cultures were treated in duplicate with 27 incremental doses of IFN-β to generate a dose-response curve (Figure 4B). In line with the increased mRNA expression (Figure 4A), there was an increase in CCL5 released in the supernatant. As expected, Figure 4C demonstrates a clear correlation between mRNA and protein expression.
Flow cytometry
Flow cytometry is a powerful tool for simultaneously investigating the expression of many intra- and/or extracellular markers. However, analyzing complex, highly interactive cultures by flow cytometry can be challenging due to cell damage and death when taking cells from culture to a single-cell suspension for processing and analysis. Comparing a variety of protocols using 0.05%-0.5% trypsin-ethylenediaminetetraacetic acid (EDTA), 1x-10x trypsin without EDTA, and gentle dissociation reagent, it was found that after treating the 6-well plate cultures with 0.05% trypsin-EDTA for 10 min at 37 °C with gentle agitation, the cells were lifted with gentle trituration to create a single-cell suspension. Especially for flow cytometric analysis of CNS cells, it is critical to be gentle during cell preparation, minimize wash steps, and take great care to keep the cells at 4 °C or on ice at all times.
To assess the viability and cell types in this single-cell suspension, cells were stained with a viability dye and fluorescently labeled antibodies against CD45, CD11b, O4, ACSA-2, and CD24 to allow visualization of each individual cell population in the cultures8 (Figure 5). Around 70% cell viability was obtained from this method, averaging approximately 2 x 106 viable, single cells per 6-well plate (Figure 5C). In line with the microscopy results (Figure 2N), there were large numbers of microglia, neurons, and astrocytes, while oligodendrocytes were the least abundant.
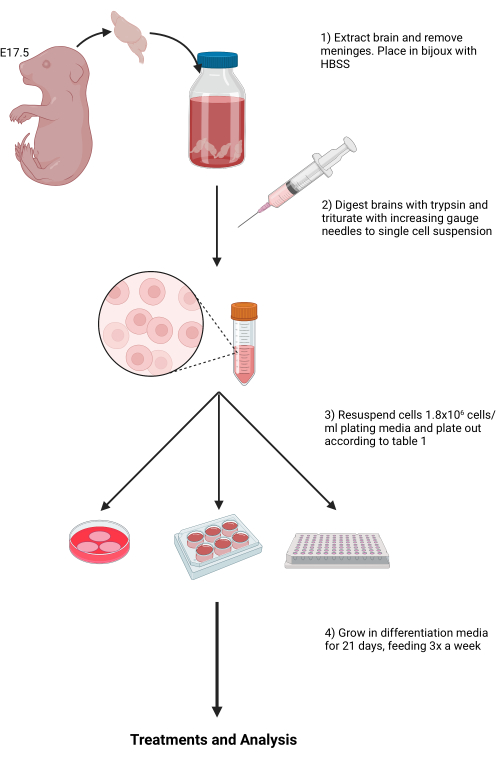
Figure 1: Schematic overview of the described method to generate mixed neuronal and glial cultures. Please click here to view a larger version of this figure.
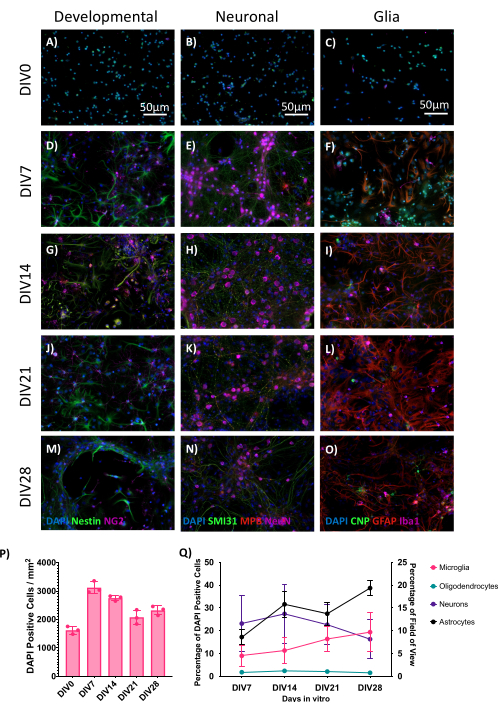
Figure 2: E17 CNS cultures stained for immunofluorescence imaging cell markers over time. Cells were fixed with 4% PFA before being permeabilized and stained to visualize different populations. Representative images of (A,D,G,J,M) developmental markers NG2 (oligodendrocyte precursor cells) and nestin (neuronal and glial precursor cells), (B,E,H,K,N) neuronal markers SMI31 (axons), MBP (myelin), and NeuN (neuron cell body), and (C,F,I,L,O) glial markers CNP (oligodendrocytes), GFAP (astrocytes), and Iba1 (microglia). Scale bars = 50 µm. (P) Counts of DAPI per mm2, n = 3, each n from technical triplicates of 10 images per triplicate. (Q) Cell counts as a percentage of DAPI+ cells (microglia, oligodendrocytes, and neurons) and percentage of the field of view (astrocytes), n = 3, each n from technical triplicates of 10 images per triplicate. Error bars are ± standard error of the mean (SEM). Please click here to view a larger version of this figure.
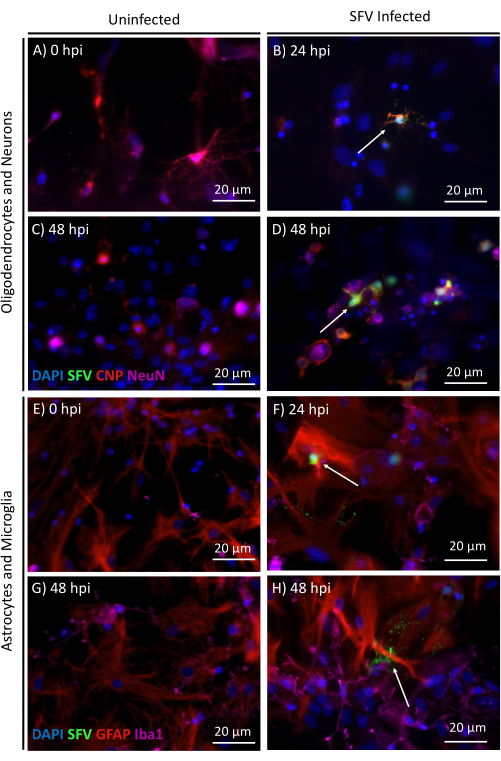
Figure 3: Semliki Forest virus (SFV) infection of E17 CNS cultures over time. Representative images of uninfected control cultures (A,C,E,G) and cultures infected with 0.05 MOI SFV (B,D,F,H). Cells were infected for 0-48 h and stained with CNP (oligodendrocyte marker) and NeuN (neuron marker) (A–D) or GFAP (astrocyte marker) and Iba1 (microglial marker) (E–H). White arrows indicate an infected cell. This strain of SFV expresses the green fluorescent protein zsGreen to enable the tracing of viral infection. Scale bars = 20 µm. Please click here to view a larger version of this figure.
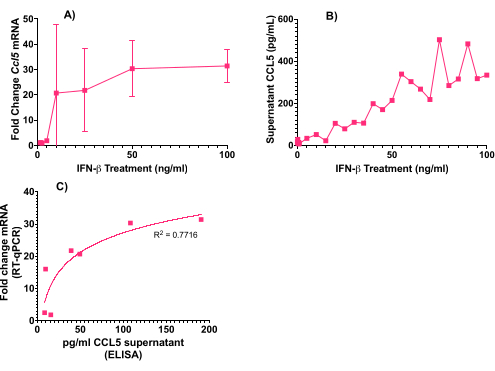
Figure 4: CCL5 mRNA and protein expression of E17 CNS cultures following IFN-β treatment. Cells were treated in 6-well plate format (mRNA) or 96-well plate format (protein) for 24 h with a range of doses of IFN-β. (A) Expression of Ccl5 mRNA after treatment with 0-100 ng/mL IFN-β, biological n = 2, each taken from technical triplicates. (B) CCL5 concentration in the supernatant following 0-100 ng/mL IFNβ treatment, biological n = 1, taken from technical triplicates. (C) Cellular mRNA Ccl5 upregulation versus CCL5 protein concentration in the supernatant with log trendline. Error bars are ± SEM. Please click here to view a larger version of this figure.
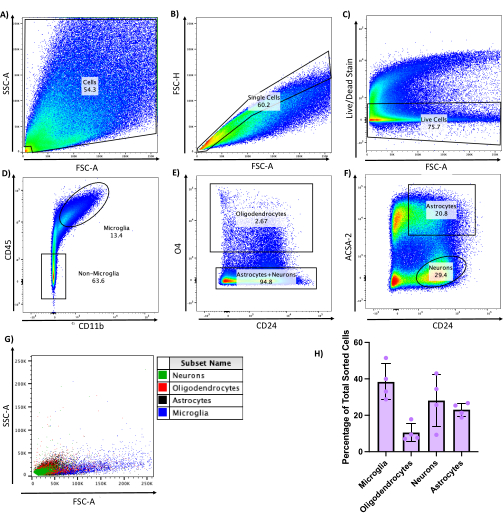
Figure 5: Flow cytometry gating strategy to generate single-cell populations from E17 CNS cultures. Cells were dissociated using 0.05% trypsin-EDTA, stained with appropriate antibodies, and sorted into individual cell populations. (A–C) Gating strategy to sort for live, single cells. (D) Strategy to select for microglia (CD45+ CD11b+). (E) Strategy to select oligodendrocytes (CD45– CD11b– O4+). (F) Strategy to sort for astrocytes (CD45– CD11b– O4– ACSA-2+ CD24+) and neurons (CD45– CD11b– O4– ACSA-2– CD24+). (G) Individual cell populations overlaid on each other, showing discrete populations. (H) Individual cell types as a percentage of the total cells sorted, n = 4. Error bars are ± SEM. Please click here to view a larger version of this figure.
| Required format | BA-PLL/well | Water for wash | Volume cells in Plating Media (PM) to plate out | Topping up Media | Remove/add when feeding cultures (3x/week) |
| 6 well format (9.6cm2) | 1000 µL | 1000 µL | 1000 µL | -400 µL | -500 µL Media |
| +600 µL DM+ | +600 µL DM+/- | ||||
| 96 well format (0.32 cm2) | 100µL | 100 µL | 50 µL | +60 µL DM+ | -50 µL Media |
| +60 µL DM+/- | |||||
| Coverslip in dish format | 20 mL per 200 coverslips | 20 mL | 100 µL per coverslip | +600 µL DM+ | -500 µL Media |
| +300 µL PM | +600 µL DM+/- |
Table 1: Required volumes for preparation of coated tissue culture plastics.
Discussion
The CNS is a complex network that spans from the brain down to the spinal cord and consists of many cell types, predominantly neurons, oligodendrocytes, astrocytes, and microglia1. As each cell has an important role in maintaining homeostasis and generating appropriate responses to challenges in the CNS9,10,11, a culture system that contains all these cell types is a useful and versatile tool to investigate how the brain might react to a stimulus. The ability to study these cells, and their interactions, in an in vitro context means a great variety of techniques can be employed to investigate various experimental questions easily. We have outlined a quick and efficient method to obtain and culture the main CNS cell types from the prenatal brain, which can mimic individual cell populations in the adult mouse brain12.
Previously established protocols for generating CNS cultures use E13.5 spinal cords4,5. In addition to having a suitable model for the spinal cord, it is highly relevant to have a model that recapitulates the brain. Therefore, brains from these E13.5 mice were originally used to generate the CNS cultures described here using the same protocol. However, after DIV14, the cells started to form large aggregates with densely packed neuronal cell bodies. While astrocytes were present, it is not clear if they were evenly distributed. It is unclear if the cells in the center of these cell bodies would receive sufficient nutrients and oxygen from the culture medium as the cells outside would. When experiencing oxygen and nutrient deprivation, neurons are liable to go through apoptosis and release stress signals to glial cells13, which might lead to a proinflammatory phenotype14. Neurogenesis was thought to be a likely cause of these densely packed aggregates, and this phase subsides by E1715. When brains from E17 mouse embryos were used, the cells still formed networks but did not cluster into the large aggregates seen in E13.5, and were therefore regarded as a more suitable age for the brain cultures to be generated from.
The current technique also results in a greater number of cells obtained from one pregnancy (averaging 1 x 107 cells/brain compared to 3-4 x 106 cells/spinal cord)7. For an average pregnant mouse with 8-10 embryos, this correlates to up to 54 different experimental conditions in the 6-well plate format, or 150 experimental conditions for microscopy (compared to 19 and 64, respectively, from the spinal cord). As such, these CNS cultures are a great tool to reduce the required number of mice16. In particular, the 96-well format can be easily used for high throughput assays, such as enabling the screening of drug candidates prior to testing in animals, vastly reducing the number of required animals for such studies. This will hopefully improve the low success rate of testing CNS drugs in animals and improve translation to clinical trials in humans17,18.
The cultures reach peak maturity by DIV21. Cell counts increase until this timepoint, after which the cells start degenerating with decreasing numbers of each type of cell. This was measured both by looking at the quantified cell numbers (Figure 2N) as well as the number of pyknotic nuclei (dense DAPI stains) (Figure 2M). While the developmental markers nestin and NG2 are still expressed on DIV14 (Figure 2G) and DIV21 (Figure 2J), this is due to some of the astrocytes going through astrogliosis, which upregulates nestin19, while some of the oligodendrocytes are not fully mature and therefore still express some NG220. Based on the microscopy validation, only a small portion of the cells still express developmental markers compared to the fully mature cells by DIV21 (Figure 2I,L); therefore, the cultures are largely mature by this time. Of note, in vivo, the density of microglia is highly variable across the murine CNS. The level of microglia, as defined by Iba1 expression using microscopy, is at the high end of this density in our cultures12. The high percentage of microglia that survive the flow cytometry procedure is likely due to microglia being more robust than other CNS cells, which generally have delicate extensions, and are therefore more likely to be damaged during flow cytometry preparation.
It only takes 3 weeks following dissection until the cells are ready for experimentation, which is shorter than most human-based in vitro methods, such as organoids or induced pluripotent stem cell-derived models21. As long as in vivo research is required to ensure the safety of new therapeutics, using in vitro cell culturing methods such as ours will aid in testing toxicity and efficacy efficiently before testing in animals, reducing the requirement for animals and enhancing the translation of research from in vitro to in vivo. Hopefully, this will eventually lead to more efficient translation to human-based clinical trials as well. Ultimately, these cultures were created to allow the study of the CNS. While in vitro systems cannot fully express the complexity of the CNS, primary cell-derived cultures more accurately represent in vivo CNS properties22,23. In vitro approaches can allow for a more reductive approach to investigate the CNS in the absence of infiltrating immune cells24 and the blood-brain barrier integrity25. Removing this layer of complexity can make it easier to unravel mechanisms. As such, the present culturing system is a useful tool to answer research questions that complement in vivo animal research.
The ability to harvest, isolate, and culture CNS cells has already greatly advanced our understanding of the innate CNS16. This article demonstrates the dissection of E17 mouse brains and the resulting trituration and culturing of the cells to generate a cell culture system containing all the main cell types of the CNS. Multiple molecular techniques have been employed on these cultures, showcasing the effectiveness these cultures have for investigating the CNS.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We would like to thank members of the Edgar and Linington labs, particularly Prof. Chris Linington, Dr Diana Arseni, and Dr Katja Muecklisch, for their advice, helpful comments, and assistance with feeding the cultures while we set up these cultures. Particular thanks go to Dr Muecklisch for providing the starting points for the Cell Profiler pipelines. This work was supported by the MS Society (grant 122) and the Yuri and Lorna Chernajovsky foundation to MP; University of Glasgow funding to JC and MP; and Wellcome Trust (217093/Z/19/Z) and Medical Research Council (MRV0109721) to GJG.
Materials
| 10x Trypsin | Sigma | T4549-100ML | To digest tissue |
| 140 mm TC Dish | Fisher | 11339283 | Put 8 35 mm dishes per 1 140 mm dish |
| 15 mL Falcon | Sarstedt | 62554502 | To collect cells into pellet for resuspension in plating media |
| 18 G needle | Henke Sass Wolf | 4710012040 | For trituration of sample |
| 21 G needle | BD | 304432 | For trituration of sample |
| 23 G needle | Henke Sass Wolf | 4710006030 | For trituration of sample |
| 35 mm TC Dish | Corning | 430165 | Plate out 3 PLL coated coverslips per 1 35 mm dish |
| 5 mL syringe | Fisher | 15869152 | For trituration of sample |
| 6 well plate | Corning | 3516 | To plate out cells for RT-qPCR, and flow cytometry |
| 7 mL Bijoux | Fisher | DIS080010R | To put brains intp |
| 96 well plate | Corning | 3596 | To plate out cells for high-throughput testing |
| ACSA-2 Antibody, anti-mouse, PE | Miltenyi | 130-123-284 | For flow cytometry staining of astrocytes |
| Angled forceps | Dumont | 0108-5/45-PO | For dissection |
| Biotin | Sigma | B4501 | For DM+/- |
| Boric Acid | Sigma | B6768-500G | For boric acid buffer |
| Brilliant Violet 421 anti-mouse CD24 Antibody, clone M1-69 | Biolegend | 101825 | For flow cytometry staining of neurons and astrocytes |
| Brilliant Violet 605 anti-mouse CD45 Antibody, clone 30-F11 | Biolegend | 103139 | For flow cytometry staining of microglia |
| Brilliant Violet 785 anti-mouse/human CD11b Antibody, clone M1/70 | Biolegend | 101243 | For flow cytometry staining of microglia |
| BSA Fraction V | Sigma | A3059-10G | For SD Inhibitor |
| CNP | Abcam | AB6319 | Mature oligodendrocytes |
| Coverslip | VWR | 631-0149 | To plate out cells for microscopy |
| Dissection Scissors | Sigma | S3146-1EA | For dissection |
| DMEM High glucose, sodium pyruvate, L-Glutamine | Gibco | 21969-035 | For DM+/-, and for plating media |
| DNase I | Thermofisher | 18047019 | For SD Inhibitor, can use this or the other Dnase from sigma |
| DNase I | Sigma | D4263 | For SD Inhibitor, can use this or the other Dnase from thermofisher |
| eBioscience Fixable Viability Dye eFluor 780 | Thermofisher | 65-0865-14 | Live / Dead stain |
| Fine forceps | Dumont | 0102-SS135-PO | For dissection |
| GFAP | Invitrogen | 13-0300 | Astrocytes |
| HBSS w Ca Mg | Sigma | H9269-500ML | For plating media |
| HBSS w/o Ca Mg | Sigma | H9394-500ML | For brains to be added to |
| Horse Serum | Gibco | 26050-070 | For plating media |
| Hydrocortisone | Sigma | H0396 | For DM+/- |
| Iba1 | Alpha-Laboratories | 019-1971 | Microglia |
| Insulin | Sigma | I1882 | For DM+ |
| Leibovitz L-15 | GIbco | 11415-049 | For SD Inhibitor |
| MBP | Bio-Rad | MCA409S | Myelin |
| Mouse CCL5/RANTES DuoSet ELISA Kit | BioTechne | DY478-05 | ELISA kit for quantifying concentration of CCL5 in supernatants of 96 well plate |
| N1 media supplement | Sigma | N6530-5ML | For DM+/- |
| Nestin | Merck | MAB353 | Neuronal stem/progenitor cells |
| NeuN | Thermofisher | PA578499 | Neuronal cell body |
| NG2 | Sigma | AB5320 | Immature oligodendrocytes |
| O4 Antibody, anti-human/mouse/rat, APC | Miltenyi | 130-119-155 | For flow cytometry staining of oligodendrocytes |
| Pen/Strep | Sigma | P0781-100ML | For DM+/-, and for plating media |
| Poly-L-Lysinehydrobromide | Sigma | P1274 | For Boric acid / poly-L-lysine solution to coat coverslips |
| SMI31 | BioLegend | 801601 | Axons |
| Sodium Tetraborate | Sigma | 221732-100G | For boric acid buffer |
| Trizol | Thermofisher | 15596026 | For lysing cells for RT-qPCR |
| Trypsin inhibitor from soybean | Sigma | T9003-100MG | For SD Inhibitor |
References
- Kovacs, G. G. Cellular reactions of the central nervous system. Handbook of Clinical Neurology. 145, 13-23 (2018).
- Russell, W. M. S., Burch, R. L. . The Principles of Humane Experimental Techniques. 1, (1959).
- Hubrecht, R. C., Carter, E. The 3Rs and humane experimental technique: Implementing change. Animals. 9 (10), 754 (2019).
- Thomson, C. E., et al. synapsing cultures of murine spinal cord-validation as an in vitro model of the central nervous system. The European Journal of Neuroscience. 28 (8), 1518-1535 (2008).
- Bijland, S., et al. An in vitro model for studying CNS white matter: Functional properties and experimental approaches. F1000Research. 8, 117 (2019).
- Fragkoudis, R., et al. Neurons and oligodendrocytes in the mouse brain differ in their ability to replicate Semliki Forest virus. Journal of Neurovirology. 15 (1), 57-70 (2009).
- Hayden, L., et al. Lipid-specific IgMs induce antiviral responses in the CNS: Implications for progressive multifocal leukoencephalopathy in multiple sclerosis. Acta Neuropathologica Communications. 8 (1), 135 (2020).
- Kantzer, C. G., et al. Anti-ACSA-2 defines a novel monoclonal antibody for prospective isolation of living neonatal and adult astrocytes. Glia. 65 (6), 990-1004 (2017).
- Yin, J., Valin, K. L., Dixon, M. L., Leavenworth, J. W. The role of microglia and macrophages in CNS homeostasis, autoimmunity, and cancer. Journal of Immunology Research. 2017, (2017).
- Gee, J. R., Keller, J. N. Astrocytes: Regulation of brain homeostasis via apolipoprotein E. The International Journal of Biochemistry and Cell Biology. 37 (6), 1145-1150 (2005).
- Madeira, M. M., Hage, Z., Tsirka, S. E. Beyond myelination: possible roles of the immune proteasome in oligodendroglial homeostasis and dysfunction. Frontiers in Neuroscience. 16, 867357 (2022).
- Keller, D., Erö, C., Markram, H. Cell densities in the mouse brain: A systematic review. Frontiers in Neuroanatomy. 12, 83 (2018).
- Wang, Y. X., et al. Oxygenglucose deprivation enhancement of cell death/apoptosis in PC12 cells and hippocampal neurons correlates with changes in neuronal excitatory amino acid neurotransmitter signaling and potassium currents. Neuroreport. 27 (8), 617-626 (2016).
- Rock, K. L., Kono, H. The inflammatory response to cell death. Annual Review of Pathology: Mechanisms of Disease. 3, 99-126 (2008).
- Chen, V. S., et al. Histology atlas of the developing prenatal and postnatal mouse central nervous system, with emphasis on prenatal days E7.5 to E18.5. Toxicologic Pathology. 45 (6), 705-744 (2017).
- Gordon, J., Amini, S., White, M. K. General overview of neuronal cell culture. Methods in Molecular Biology. 1078, 1-8 (2013).
- Kesselheim, A. S., Hwang, T. J., Franklin, J. M. Two decades of new drug development for central nervous system disorders. Nature Reviews Drug Discovery. 14 (12), 815-816 (2015).
- Gribkoff, V. K., Kaczmarek, L. K. The need for new approaches in CNS drug discovery: Why drugs have failed, and what can be done to improve outcomes. Neuropharmacology. 120, 11-19 (2017).
- Zamanian, J. L., et al. Genomic analysis of reactive astrogliosis. Journal of Neuroscience. 32 (18), 6391-6410 (2012).
- Nishiyama, A., Suzuki, R., Zhu, X. NG2 cells (polydendrocytes) in brain physiology and repair. Frontiers in Neuroscience. 8, 133 (2014).
- Lee, C. T., Bendriem, R. M., Wu, W. W., Shen, R. F. 3D brain organoids derived from pluripotent stem cells: Promising experimental models for brain development and neurodegenerative disorders. Journal of Biomedical Science. 24 (1), 59 (2017).
- Asch, B. B., Medina, D., Brinkley, B. R. Microtubules and actin-containing filaments of normal, preneoplastic, and neoplastic mouse mammary epithelial cells. Cancer Research. 39 (3), 893-907 (1979).
- Koch, K. S., Leffertt, H. L. Growth control of differentiated adult rat hepatocytes in primary culture. Annals of the New York Academy of Sciences. 349, 111-127 (1980).
- Nevalainen, T., Autio, A., Hurme, M. Composition of the infiltrating immune cells in the brain of healthy individuals: effect of aging. Immunity and Ageing. 19 (1), 45 (2022).
- Daneman, R., Prat, A. The blood-brain barrier. Cold Spring Harbor Perspectives in Biology. 7 (1), 020412 (2015).

