A Peg Plate Biofilm Assay for Screening Antibacterial Agents
Abstract
Source: Dalecki, A. G., et al. Targeting Biofilm Associated Staphylococcus aureus Using Resazurin Based Drug-susceptibility Assay. J. Vis. Exp. (2016)
This video demonstrates an assay for screening antimicrobial compounds that efficiently eliminate biofilms using a peg-plate. The methodology monitors the biofilm's metabolic state after treatment with antimicrobial compounds, facilitating the determination of the minimum biofilm eradication concentration.
Protocol
1. Biofilm Initiation
- Grow a culture of biofilm-producing organisms in a nutrient-rich medium. Inoculate Staphylococcus aureus strain Newman in 10 ml of Mueller-Hinton medium from a glycerol stock. Perform all work involving handling of S. aureus with gloves and within a biosafety cabinet.
- Incubate for 16 hr at 37 °C on a rotary shaker (100-200 rpm).
- Determine OD600 of the culture using a spectrophotometer and a standard cuvette (path length: 1cm).
- Calculate the volume (V) of culture needed to prepare 20 ml of inoculum with OD600(inoculum) of 0.1 in chelexed Roswell Park Memorial Institute media (CRPMI) using the formula: [V = 20 ml*0.1/OD600(culture)].
- Add the calculated culture volume (from above) to a centrifuge tube and pellet cells by spinning for 1 min at 10,000 x g. To prepare biofilm inoculum, aspirate the spent growth medium and resuspend the cell pellet in 20 ml CRPMI.
- Pour inoculum into a 25 ml reservoir.
- Carefully remove the lid from a sterile 96-well peg plate by pulling it straight up.
Note: Care must be taken not to contaminate the pegs hanging down from the lid. The lid can be placed upside down on a clean surface within a biosafety cabinet. - Using a 200 µl multi-channel pipette, add 125 µl inoculum to each well of rows A to G of the bottom plate.
- Fill 125 µl sterile CRPMI medium in each well of row H as a sterility control.
- Take the peg lid and hold it above the plate bottom with the pegs facing downwards. Carefully align the individual pegs with their respective wells. Avoid physical contact between the plate and the pegs. Once aligned, lower the lid carefully down. Avoid misalignments as readjustments performed after the pegs have touched the bottom plate may contaminate sterility controls in row H.
- Seal the lid to the plate with paraffin film to prevent evaporation and place it in a sealable plastic bag, sealing tightly. Keep the air volume in the bag as low as possible to minimize evaporation.
- Incubate without shaking at 37 °C for 48 hr in a 5% CO2 incubator.
Note: 48 hr of growth has been optimal for S. aureus, but will vary based upon the organism used.
Note: Although static incubation is a good starting point to optimize the assay, gently shaking at 150 rpm on a microplate shaker is a possible variation that may enhance the biofilm formation of some S. aureus isolates.
2. Preparation of the Biofilm Challenge Plate
- On the day of the biofilm challenge take a fresh 96-well plate and add 100 µl of CRPMI, equilibrated to RT, to each well of rows B to H. To avoid inconsistent results, utilize a 96-well plate that can hold 200 µl of media when the pegs are inside, and features wells with dimensions identical to those of the biofilm initiation plate.
- Dilute up to 4 test compounds separately in 1.0 ml of CRPMI to 2x the desired final concentration, in order to account for a final dilution in step 2.7.
Note: A good starting point is 8 fold above the inhibitory concentration of planktonic cells. - Add 200 µl of compound 1 in wells A1 to A3, compound 2 in wells A4 to A6, compound 3 in wells A7 to A9, and compound 4 in wells A10 to A12.
- Serially dilute the test compounds by using a multichannel pipette and transfer 100 µl from row A to row B and mix.
- In that manner, continue to transfer 100 µl well to well. Mix after each transfer and stop in row F.
- After mixing, expel 100 µl from row F into a waste container. Do not transfer any liquid to rows G and H.
- Add 100 µl of CRPMI to each well of the plate using a multichannel pipette to bring the volume to 200 µl. See Figure 1 for the final plate layout.
3. Biofilm Challenge
- Add 200 µl of phosphate-buffered saline (PBS) to a fresh sterile 96-well plate that features wells with dimensions identical to those of the biofilm initiation plate.
- Remove the plastic bag and paraffin film from the incubated biofilm initiation plate.
- Lift the lid straight up. Avoid contact between the pegs and the plate bottom as this may damage the biofilm. Keep the bottom part for subsequent OD600 analysis (see 3.8).
- Rinse off planktonic cells by placing the peg-lid onto the plate containing PBS (from step 3.1). Submerge pegs for 5 sec, lift out of PBS, and submerge once more, being careful not to hit the pegs against the well sides.
- Place the rinsed peg lid into the challenge plate. Avoid unnecessary contact between pegs and bottom plate as this will damage the biofilm leading to inconsistent results.
- Seal the plate with paraffin film and place it into a sealable plastic bag as described in step 1.10.
- Incubate the plate without shaking for 24 hr at 37 °C in a 5% CO2 incubator.
- Take the bottom of the biofilm initiation plate from 3.3. Resuspend the bacteria in each well using a multichannel pipette and measure the OD600 with a suitable microplate reader to confirm growth occurring in all wells but the sterility controls in row H.
4. Biofilm Determination
- Use a plate reader equipped with an incubation chamber and capable of taking kinetic fluorescence readings for detecting resazurin conversion. Set up a 24-hour kinetic fluorescence measurement using a 530 nm excitation and 590 nm emission.
- Set the chamber temperature to 37 °C and have the plate read every 20 min. Set the plate reader to measure from the bottom of the plate according to the manufacturer's instructions.
- Prepare the wash plate by adding 200 µl of PBS with a multichannel pipette to a sterile 96-well plate.
- Prepare biofilm recovery medium by adding 400 µl of 0.8 mg/ml resazurin stock solution to 20 ml of CRPMI medium to a final concentration of 16 µg/ml resazurin.
Note: Resazurin is a known skin irritant. Use a lab coat, splash goggles, and gloves while handling. CRPMI as the biofilm recovery media was used because it provides the lowest background signal, but it can be replaced by Mueller Hinton media if appropriate. - Add 150 µl of biofilm recovery medium with a multichannel pipette to each well of a fresh 96-well plate.
- Transfer the challenge plate from the incubator into a biosafety cabinet and carefully remove the plastic bag and sealing film.
- Remove the peg lid from the challenge plate and rinse off planktonic cells in PBS as described in 3.4. Keep the bottom of the challenge plate for subsequent OD600 analysis (see 4.10).
- After rinsing, place the peg lid into the plate bottom containing the biofilm recovery medium.
- Wrap the side of the plate with paraffin film, being careful not to obstruct the bottom of the perimeter wells. Immediately after wrapping the plate, place it on the plate reader and start a kinetic read over a 24-hour period by initiating the previously made resazurin kinetic protocol from step 4.1.
- To quantify the growth of planktonic cells that may or may not occur during the challenge, read the OD600 of the entire challenge plate bottom using a microplate reader. Follow the instructions given in step 3.8.
Note: Carefully resuspend the content of each well as cells shedding off the biofilm tend to grow in clumps at the bottom of the well. Any bubbles within the well will strongly interfere with OD600 readings and must be avoided or removed prior to the read.
Representative Results
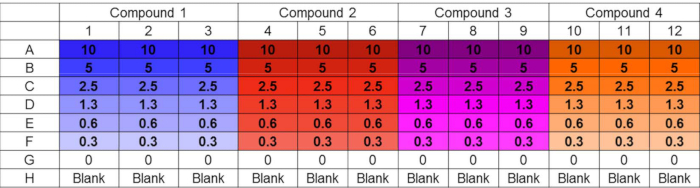
Figure 1. Challenge plate layout. Example layout of challenge plate accommodating parallel testing of four compounds in triplicate at six distinct concentrations. Untreated and sterility controls are also included. The layout design supports convenient in-plate preparation of serial dilutions using a multichannel pipet. Concentrations are in µM.
Declarações
The authors have nothing to disclose.
Materials
| Mueller-Hinton Medium | Oxoid | CM0405 | Follow recommendations of manufacturer |
| RPMI-medium | Corning | 17-105-CV | |
| CRPMI | Ref 9 | RPMI-1640 medium chelexed for 1 hr with Chelex 100 resin and then supplemented with 10% unchelexed RPMI-1640 | |
| Chelex 100 Resin | Bio Rad | 142-2822 | |
| MBEC-plates | Innovotech | 19111 | |
| Resazurin Sodium Salt | Sigma | R7017 | 800 µg/ml in DI water Filter sterile |
| Micro Plate Shaking Platform | Heidolph Titramax 1000 | ||
| Cytation 3 Plate Reader | Biotek | ||
| Gen5 software | Biotek | Recording and analysis of resazurin conversion | |
| Neocuproine | Sigma | N1501 | Prepare 10 mM stock in 100% Ethanol, store at -80 ºC |
| Copper sulfate | Acros Organics | 7758-99-8 | Prepare a 100 mM stock solution in water, store at 4 ºC |
| Cu-Neocuproine | Self-Made | Generated by mixing equal molarities of neocuproine and copper sulfate. Mix was diluted in CRPMI medium to desired concentration. | |
| Gentamicin | Sigma | G3632-1G |

