A Peptide Array-Based Detection of Anti-donor HLA Alloantibodies in Organ Recipients
Abstract
Source: Liu, P., et al. Personalized Peptide Arrays for Detection of HLA Alloantibodies in Organ Transplantation. J. Vis. Exp. (2017).
This video demonstrates an assay to detect anti-donor HLA alloantibodies in organ recipients. A peptide array is synthesized to represent potential epitopes for antibody-mediated organ rejection. The array is incubated with the recipient's serum, containing alloantibodies that bind to their target epitopes. Antibody binding is detected using chemiluminescence to identify donor-specific HLA epitopes that incite alloantibody reactivity.
Protocol
All procedures involving sample collection have been performed in accordance with the institute's IRB guidelines.
1. Design of Custom Array Layout and Production
- Generate a spreadsheet of peptide sequences with corresponding allele callings. The array has 20 rows and 30 columns and can hold up to 20×30=600 spots for peptides. Depending on the total number of peptides recorded in step 1.5 from one particular donor, and based on our past experience with two examples that we had tried, the 600 spot array can hold all the sequences generated from ~2 separate transplant cases.
- Rationally plan the most efficient way to fit the sets of peptides within the 600-spot format of the array.
- If more than one subject can fit into one whole array, insert empty rows after the first donor's sequences to match the total number of rows that can be divided by 30 (the number of spots in a row on the array).
- Immediately following the last empty row for the first set of sequences, paste in the entire column of sequence contents from the second transplant case.
- Repeat these steps until the array is filled and no more cases can be added without exceeding the 600 spots. If entire empty rows are left to fill at the bottom, "move" the empty row to be positioned between the two donor sets. This wide space left between cases makes cutting of the membrane after completion of the synthesis easier – following step 2.2. below.
- Program the peptide sequences in the context of the array layout. The program of the SPOT synthesizer takes a text format of the sequences (one row, one peptide sequence), which can be directly obtained by saving the resulting spreadsheet from step 1.1.4 as a simple text document (without the extra column(s) for allele names, aa positions etc.).
- Run automated peptide array synthesis via the SPOT synthesizer. The entire operation of the synthesis is detailed in Kudithipudi et al. Note that Fmoc synthesis of two arrays takes ~4-5 days.
2. Probe and Reprobe Antisera from a Time Series of an Individual Transplant Recipient.
NOTE: The 600-spot membrane array has the dimensions ~7 cm x 13 cm. After synthesis, the arrays can be stored at room temperature as dry membranes for at least two years when shielded from direct light. Avoid excessive folding of the membrane to preserve its longevity for repeated use.
- Rehydrate the membrane array with ethanol and visualize peptide spots stained with Ponceau S.
- Rehydrate the membrane array carrying the peptides following a stepwise procedure optimized in Li et al.
- Immerse the array membrane in 20 mL of 100% ethanol.
- Add 20 mL distilled water to dilute the solution to 50% ethanol and incubate at room temperature for 15 min.
- Change the immersion solution to 40 mL of 100% water three times and incubate 15 min each time.
- Wash in a proper working buffer, e.g., 20 mL of TBST (Tris-buffered saline with 0.1%), three times for 5 min each.
- Ponceau S staining of the array to visualize synthetic peptides
- Add 20 mL of pre-formulated Ponceau S solution directly to the hydrated array and incubate for ~30 s with shaking.
- Run distilled water continuously over the membrane to de-stain the background Ponceau S color. During the process, peptide spots of red color may become visible.
- Separate the portions of the array for each donor by carefully cutting between the sections of the array for individual cases. Mark the orientation of each array.
- Rehydrate the membrane array carrying the peptides following a stepwise procedure optimized in Li et al.
- Pre-block array.
- Block the membrane in 20 mL of 5% non-fat milk dissolved in TBST buffer. This milk-based solution will further de-stain the Ponceau S color. Replace the milk buffer several times in order to achieve the clearest peptide images.
- For record-keeping purposes, take a photo of the Ponceau S image of the array using a hand-held camera (example in Figure 1).
- Continue blocking of the membrane in 5% non-fat milk at 4 °C overnight or at room temperature for 2 h with rocking.
- Incubate array with transplant antiserum.
NOTE: It should be noted that a phenomenon known as prozone or the Hook effect may result from complement-dependent interference in serum antibody analysis. To circumvent this problem, an optional serum pretreatment step can be considered with either ethylenediaminetetraacetic acid (EDTA) or heat inactivation of serum samples.- Remove the blocking buffer and wash the membrane with 20 mL of TBST three times the next day for 5 min each time.
- Add 20 µL of the crude serum of the recipient to 20 mL of 2.5% milk in TBST buffer and incubate with the membrane for 2-3 h at room temperature. Note that in this initial round of probing, it is recommended that the last post-transplant serum (or likely most sensitized serum) in the time series is first used. This is based on the assumption that earlier specimens in the series, particularly from pre-transplant time points, have less variety of alloantibodies that tend to develop over time. This way, any possibility of signal interference from "carry-over" between probing rounds can be clearly distinguished from truly developed alloantibody reactivity due to immune responses against the graft.
- Wash the array using 20 mL of TBST and incubate it with a secondary antibody.
- Wash the membrane three times for 10 min each time.
- Incubate with goat anti-human IgG-HRP (Horseradish peroxidase) secondary antibody at 1:10,000 dilution in TBST buffer supplemented with 1% milk for another 2 h.
- Wash and develop the blot.
- Wash the membrane three times with TBST for 10 min each time.
- Perform enhanced chemiluminescence (ECL) using 5 mL of luminol solution freshly mixed with 5 mL of peroxide solution to develop the membrane (for 1 min).
- Visualize ECL signals using a suitable imager (i.e., ChemiDoc Imaging Systems or Azure C600).
- Scan and quantify the blot.
- Save the developed images (Figure 2, lower image) and perform quantification of spot intensity.
3. Compare Antiserum Reactivity across a Clinical Time Series.
- Strip the array.
- At this point, keep the membrane wet at all times.
- Strip the membrane by incubating with 20 mL of commercial stripping buffer at 37 °C for 20 min, and then wash the membrane with TBST three times for 10 min each. Then repeat the blocking (step 3.2) and probing (step 3.3) steps using a different serum of the patient taken at a different time point.
- Block stripped membrane.
Note: Following stripping, the membrane can be reused for another round of probing of a different serum from the same patient in a time series.- Block the membrane using 5% milk buffer as before (see step 2.2).
- Reprobe a different antiserum from the same time series (repeat steps from 2.3-2.6; example in Figure 2, upper image).
NOTE: The stripped array can then be used to reprobe another serum specimen. Since the peptides are covalently conjugated to the supporting matrix of the membrane, we showed that the array can be reused for up to 20 rounds of stripping and reprobing cycles without losing its performance.
Representative Results
Figure 1. Image of an array section stained with Ponceau S
Note disparity in color intensity is due to differences in amino acid composition among peptides, while peptide concentrations among all spot areas are the same. (The image is an illustrative example, not the one from this actual study.)
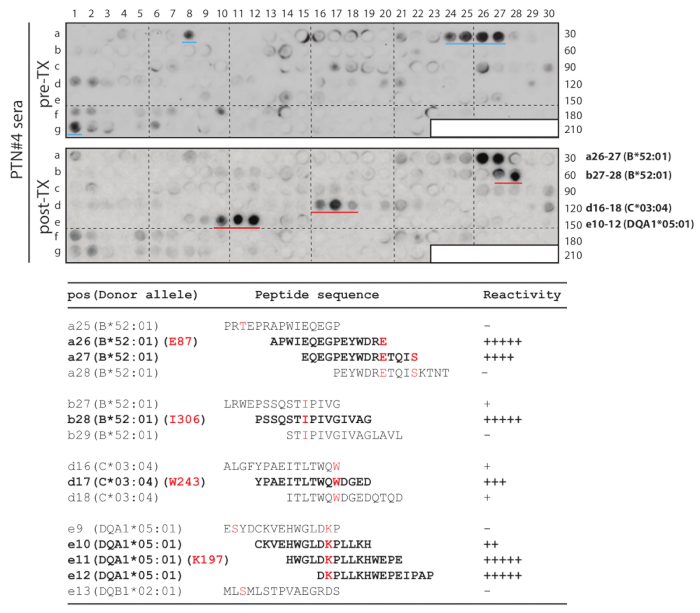
Figure 2. Donor-specific HLA-A, B, C, DQ, DR array study of mismatched epitopes in PTN#4.
Serial peptides were derived from the donor's sequences to cover mismatched residues (An example of DQB1 in Figure 3). The array was used to probe the post-transplant serum (lower blot: post-TX) and, subsequently, the pre-transplant serum (upper blot: pre-TX) from the same patient. The four sets of strong spots from post-transplant probing are marked by red lines (in the lower blot), while two medium-intensity spots that were only associated with pre-transplant serum are marked by blue lines (in the upper blot). The bottom table shows the corresponding peptide sequences and their reactivity to the post-transplant serum. Donor-specific (mismatched) residues E87, I306, W243 and K197 of their respective alleles are in red letters, and peptides showing strong antibody reactivity are in bold fonts. This figure was adapted and modified by Liu et al.
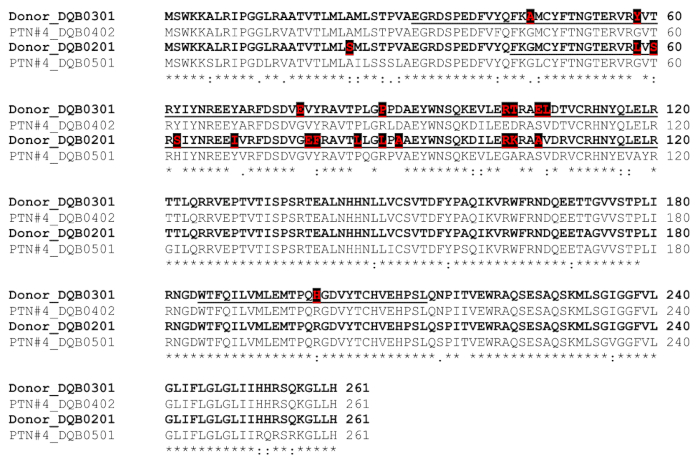
Figure 3. Comparison of donor vs. patient/recipient HLA DQB1 and selection of template sequences for peptide synthesis.
The donor's sequences are in bold, with donor-specific mismatches in red font and also highlighted with black boxes. Template sequences (underlined) for deriving peptides contain these donor-specific residues. (This figure was adapted and modified from Liu et al.)
Declarações
The authors have nothing to disclose.
Materials
| Peptide array | INTAVIS Bioanalytical Instruments | ||
| Ethanol | Sigma-Aldrich | E7023 | |
| Ponceau S solution | Sigma-Aldrich | P7170 | |
| Non-fat milk | Bio Rad Laboratories | 1706404 | |
| TBST | Santa Cruz Biotechnology | 10711454001 | |
| Goat anti-human IgG–HRP | ThermoFisher Scientific | A18811 | |
| Clarity Western ECL Substrate | Bio Rad Laboratories | 1705061 | |
| Restore Western Blot Stripping Buffer | Thermo Scientifics | 21059 | |
| ChemiDoc gel imaging system | Bio Rad Laboratories | 1708265 |

