Modified Annexin V/Propidium Iodide Apoptosis Assay For Accurate Assessment of Cell Death
Summary
An accurate method for the assessment of cell death is described. The protocol improves upon conventional Annexin V/ propidium iodide (PI) protocols, which display up to 40% false- positive events in cell lines and primary cells from a broad range of animal models.
Abstract
Studies of cellular apoptosis have been significantly impacted since the introduction of flow cytometry-based methods. Propidium iodide (PI) is widely used in conjunction with Annexin V to determine if cells are viable, apoptotic, or necrotic through differences in plasma membrane integrity and permeability1,2. The Annexin V/ PI protocol is a commonly used approach for studying apoptotic cells3. PI is used more often than other nuclear stains because it is economical, stable and a good indicator of cell viability, based on its capacity to exclude dye in living cells 4,5. The ability of PI to enter a cell is dependent upon the permeability of the membrane; PI does not stain live or early apoptotic cells due to the presence of an intact plasma membrane 1,2,6. In late apoptotic and necrotic cells, the integrity of the plasma and nuclear membranes decreases7,8, allowing PI to pass through the membranes, intercalate into nucleic acids, and display red fluorescence 1,2,9. Unfortunately, we find that conventional Annexin V/ PI protocols lead to a significant number of false positive events (up to 40%), which are associated with PI staining of RNA within the cytoplasmic compartment10. Primary cells and cell lines in a broad range of animal models are affected, with large cells (nuclear: cytoplasmic ratios <0.5) showing the highest occurrence10. Herein, we demonstrate a modified Annexin V/ PI method that provides a significant improvement for assessment of cell death compared to conventional methods. This protocol takes advantage of changes in cellular permeability during cell fixing to promote entry of RNase A into cells following staining. Both the timing and concentration of RNase A have been optimized for removal of cytoplasmic RNA. The result is a significant improvement over conventional Annexin V/ PI protocols (< 5% events with cytoplasmic PI staining).
Protocol
This procedure can be performed in 500 μL siliconized tubes or 6 mL polystyrene round-bottom FACS tubes. The latter is normally used when carrying out viability analysis in conjunction with other procedures. All volumes given are for 6 mL polystyrene round-bottom FACS tubes. For 500 μL siliconized tubes, reduce all volumes by 1/5.
The optimum concentration for flow cytometry analysis is 2-4 x 106 cells per 200 μL volume. Cell loss may result from this procedure and thus we recommend that each sample consist of 4 x 106 cells at the start of the procedure.
1. Cell Preparation
- Harvest cells – refer to specific procedures for corresponding cell lines or primary cell isolations.
- Centrifuge samples at 335 x g for 10 minutes and decant the supernatant.
- Resuspend cells in 2 mL 1 x phosphate buffered saline (PBS) -/- (no calcium, no magnesium).
- Centrifuge samples at 335 x g for 10 minutes and decant the supernatant.
- Resuspend cells in 1 mL 1 x Annexin V binding buffer.
- Centrifuge samples at 335 x g for 10 minutes and decant the supernatant.
- Resuspend cells in 100 μL 1 x Annexin V binding buffer.
2. Application of Annexin V/ PI Stain
- Add Annexin V according to the manufacturer’s recommendations [e.g. 5 μL Annexin V Alexa Fluor 488 (Molecular Probes, A13201)].
- Incubate tubes in the dark for 15 minutes at room temperature.
- Add 100 μL of 1 x Annexin V binding buffer to each reaction tube. There should be approximately 200 μL in each tube.
- Add 4 μL of PI (Sigma, Cat# P-4864-10ML) that has been diluted 1:10 in 1 x Annexin V binding buffer (i.e. 1 μL PI with 9 μL 1 x Annexin V binding buffer). This will yield a final PI concentration of 2 μg/mL in each sample.
- Incubate tubes in the dark for 15 minutes at room temperature.
- Add 500 μL 1 x Annexin V binding buffer to wash the cells.
- Centrifuge samples at 335 x g for 10 minutes and decant the supernatant.
- Resuspend cells in 500 μL 1 x Annexin V binding buffer and 500 μL 2% formaldehyde to create a 1% formaldehyde (fixative) solution. Mix tubes by gentle flicking.
- Fix samples on ice for 10 minutes. Alternatively, samples can be stored overnight at 4°C in the dark. If the latter method is chosen, make sure to be consistent across all tubes labeled with Annexin V/PI (including compensation controls).
- Add 1 mL 1 x PBS-/- to each sample and mix gently by flicking.
- Centrifuge tubes at 425 x g for eight minutes and decant the supernatant.
- Repeat steps 2.10 and 2.11.
- Resuspend pellet by flicking the tube.
- Add 16 μL of 1:100 diluted RNase A (Sigma, R4642) to give a final concentration of 50 μg/mL. Incubate for 15 min at 37°C.
- Add 1 mL 1 x PBS-/- and mix gently by flicking.
- Centrifuge tubes at 425 x g for eight minutes.
- Samples are now ready to be analyzed. Alternatively, samples can be used for subsequent staining steps if Annexin/PI staining is being performed in parallel with other procedures.
3. Representative Results:
Examples of cell types and cell lines that have varying degrees of false-positive PI staining are shown in Figure 1. To account for the false-positive events, cells were fixed and then treated with RNase A. This results in a significant decrease in the number of false-positive events, compared to cells that didn’t undergo RNase treatment (Figure 2B). Figure 2C shows representative images of cells that underwent RNase treatment, compared to cells that had no RNase treatment. Figure 3 shows how the modified Annexin V/PI protocol, with fixation and RNase treatment steps, improves upon conventional protocols by limiting the number of false-positive staining events.
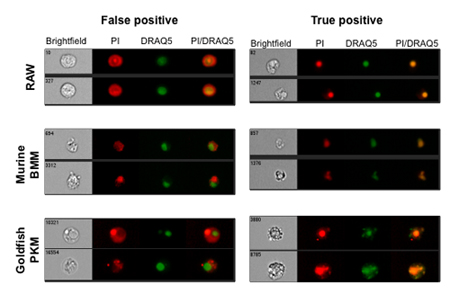
Figure 1. Conventional propidium iodide staining protocols lead to a significant number of false positive events. We have previously evaluated extent of false positive staining across primary cells across a broad range of animal models as well as in a variety of cell lines 10. Representative images show extent of false positive staining in three unique cell populations (one commonly used cell line – RAW macrophages and two primary cells – murine bone marrow macrophages (BMM) and goldfish primary kidney macrophages (PKM)). True positive events display the expected co-localization between PI and DRAQ5 within the nuclear compartment (yellow areas depict colocalization between PI and DRAQ5). DRAQ5 is highly membrane permeable dye that accurately stains nuclear compartment in both live and dead cells 10,11,12. For easier visual determination of colocalization DRAQ5 stain was given a green pseudocolour.
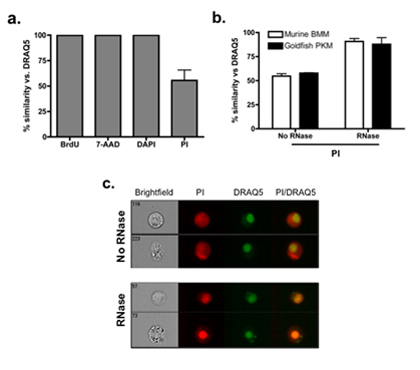
Figure 2. Improving the accuracy of PI nuclear staining. (A) We assessed accuracy of PI nuclear staining based on degree of similarity of a number of well-characterized nuclear stains (BrdU, 7-AAD and DAPI) to DRAQ5. BrdU is a synthetic nucleoside that is incorporated into the DNA of proliferating cells, and as such will only stain within the nuclear compartment. 7-AAD has a high affinity for DNA and, similar to PI, cannot cross an intact plasma membrane. DAPI also has a strong affinity for DNA and is the most commonly used nuclear stain in fluorescent microscopy. The degree of similarity was >99% between BrdU, 7-AAD, DAPI and DRAQ5, highlighting their ability to accurately stain the cell nucleus in RAW 264.7 macrophages. In contrast, cytoplasmic PI staining based on conventional protocols leads to a marked decrease in percent similarity of staining relative to DRAQ5. (B) Similar results are observed in two primary cells tested: murine bone marrow macrophages (BMM) and goldfish primary kidney macrophages (PKM). Incorporation of 50 μg/mL RNase A at specific stage within the staining procedure removes non-nuclear PI staining and significantly increases degree of similarity relative to DRAQ5 staining. (C) Representative images of primary kidney macrophages show the degree of similarity for cells with and without RNase treatment. For easier visual determination of differences between treatments, DRAQ5 was given a green. Yellow areas depict colocalization between BrdU and DRAQ5, and PI and DRAQ5.
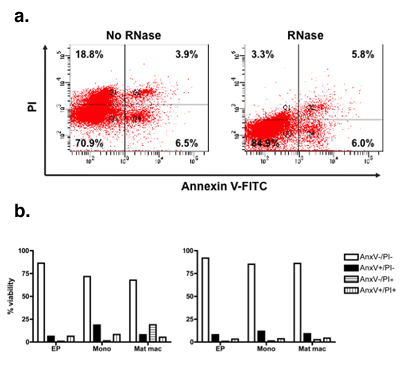
Figure 3. Modified Annexin V/PI significantly reduces false apoptosis/necrotic events. Primary goldfish kidney macrophages (PKM) were stained with conventional and modified Annexin V/PI protocols. Scatterplots depict the Annexin V/PI stain in goldfish PKM. The scatterplot on the left shows conventional Annexin V/PI staining (no RNase) of PKM cells. The scatterplot on the right shows PKM cells stained with the modified Annexin V/PI protocol (with RNase). The addition of RNase results in a marked reduction of PI staining due to the removal of false positive stains. PKM cells were then analyzed based on distinct populations within the cultures- early progenitors (EP), monocytes (Mono) and mature macrophages (Mat mac). The reduction in the number of positive PI events, due to removal of false positive events is more pronounced in large mature macrophage cells than in smaller early progenitor cells. Conclusions based on conventional protocols would have erroneously suggested a positive correlation in cellular death for more mature macrophage subsets.
Discussion
The Annexin V/PI protocol presented here is a modified version of conventional protocols and takes into account the presence of RNA in the cytoplasm which also has high affinity for PI. Introduction of RNase A (50 μg/mL) following a 1% formaldehyde fixation step late in the staining procedure significantly improves accuracy of nuclear PI staining. No negative effects are observed in nuclear PI staining or plasma membrane Annexin V staining. Without RNase A treatment, up to 40% of PI stain results can lead to false positive events, which leads to a potentially significant and negative impact on downstream conclusions10.
Our modified Annexin V/PI staining protocol is simple and has been used effectively in a broad range of cell types (Primary cells: murine bone marrow macrophages and splenocytes; swine lung resident cells, lung alveolar macrophages, mesenteric lymph node isolates, peripheral blood leukocytes, splenocytes; chicken blastoderm cells; goldfish primary kidney macrophages; carp peripheral blood leukocytes; Cell lines: RAW 264.7 macrophages, Jurkat T cells, swine 3D4/31 macrophages, CCL-71 goldfish fin fibroblasts, 3B11 catfish B cells 10). It is important to note that cells are differentially affected by false positive PI staining, with primary cells generally having a greater number of false positive events 10. The differences false positive PI staining among these cellular populations may be partially attributed to differences in cell size, but may also result from differences in RNA content. As such, incorporation of this procedure is particularly relevant for experimental systems that utilize, among others, cells undergoing genotoxic stress, cells treated with cell cycle arrest drugs such as thymidine or hydroxyurea, virally-infected cells, and studies on embryonic cells where developmental progression is characterized by discrete changes in cellular RNA synthesis.
Declarações
The authors have nothing to disclose.
Acknowledgements
This study was supported by a Natural Sciences and Engineering Council of Canada (NSERC) research grant and an Alberta Agriculture Funding Consortium grant to DRB. AMR is supported through an NSERC Vanier Canadian Graduate Scholarship, a University of Alberta teaching assistantship and a Queen Elizabeth II graduate scholarship.
Materials
| Material Name | Tipo | Company | Catalogue Number | Comment |
|---|---|---|---|---|
| 1 x PBS-/- | ||||
| 1 x Annexin V Binding Buffer | BD Biosciences | 556454 | ||
| Annexin V-Alexa Fluor 488 | Molecular Probes (Invitrogen) | A13201 | ||
| Propidium iodide (PI) | Sigma-Aldrich | P4864 | 1 mg/mL in H2O | |
| 2% Formaldehyde | Sigma-Aldrich | F1268 | ||
| RNase A from bovine pancreas | Sigma-Aldrich | R4642 | ||
| DRAQ5 | Biostatus | DR50050 | Dilute 1:60 in 1 x PBS-/- |
Referências
- Vermes, I., Haanen, C., Steffens-Nakken, H., Reutelingsperger, C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labeled Annexin V. J Immunol Methods. 184, 39-51 (1995).
- Vermes, I., Haanen, C., Reutelingsperger, C. Flow cytometry of apoptotic cell death. J Immunol Methods. 243, 167-190 (2000).
- Cornelissen, M., Philippe, J., De Sitter, S., De Ridder, L. Annexin V expression in apoptotic peripheral blood lymphocytes: An electron microscopic evaluation. Apoptosis. 7, 41-47 (2002).
- Fried, J., Perez, A. G., Clarkson, B. D. Flow cytofluorometric analysis of cell cycle distributions using propidium iodide. Properties of the method and mathematical analysis of the data. J Cell Biol. 71, 172-181 (1976).
- Bacso, Z., Everson, R. B., Eliason, J. F. The DNA of Annexin V-binding apoptotic cells is highly fragmented. Cancer Res. 60, 4623-4628 (2000).
- Darzynkiewicz, Z., Bruno, S., Del Bino, G., Gorczyca, W., Hotz, M. A., Lassota, P., Traganos, F. Features of apoptotic cells measured by flow cytometry. Cytometry. 13, 795-808 (1992).
- Kroemer, G., Dallaporta, B., Resche-Rigon, M. The mitochondrial death/life regulator in apoptosis and necrosis. Annu. Rev. Physiol. 60, 619-642 (1998).
- Denecker, G., Vercammen, D., Declercq, W., Vandenabeele, P. Apoptotic and necrotic cell death induced by death domain receptors. Cell Mol. Life Sci. 58, 356-370 (2001).
- Faleiro, L., Lazebnik, Y. Caspases disrupt the nuclear-cytoplasmic barrier. J Cell Biol. 151, 951-959 (2000).
- Rieger, A. M., Hall, B. E., Luong, L. T., Schang, L. M., Barreda, D. R. Conventional apoptosis assays using propidium iodide generate a significant number of false positives that prevent accurate assessment of cell death. J Immunol Methods. 358, 81-92 (2010).
- Edward, R. Minireview: Use of DNA-specific anthraquinone dyes to directly reveal cytoplasmic and nuclear boundaries in live and fixed cells. Mol. Cells. 27, 391-396 (2009).
- Njoh, K. L., Patterson, L. H., Zloh, M., Wiltshire, M., Fisher, J., Chappell, S., Ameer-Beg, S., Bai, Y., Matthews, D., Errington, R. J., Smith, P. J. Spectral analysis of the DNA targeting bisalkylaminoanthraquinone DRAQ5 in intact living cells. Cytometry Part A. 69, 805-814 (2006).

