Hybridization in situ of Salivary Glands, Ovaries, and Embryos of Vector Mosquitoes
Summary
Temporal and spatial gene expression analyses have a crucial role in functional genomics. Whole-mount hybridization in situ is useful for determining the localization of transcripts within tissues and subcellular compartments. Here we outline a hybridization in situ protocol with modifications for specific target tissues in mosquitoes.
Abstract
Mosquitoes are vectors for a diverse set of pathogens including arboviruses, protozoan parasites and nematodes. Investigation of transcripts and gene regulators that are expressed in tissues in which the mosquito host and pathogen interact, and in organs involved in reproduction are of great interest for strategies to reduce mosquito-borne disease transmission and disrupt egg development. A number of tools have been employed to study and validate the temporal and tissue-specific regulation of gene expression. Here, we describe protocols that have been developed to obtain spatial information, which enhances our understanding of where specific genes are expressed and their products accumulate. The protocol described has been used to validate expression and determine accumulation patterns of transcripts in tissues related to mosquito-borne pathogen transmission, such as female salivary glands, as well as subcellular compartments of ovaries and embryos, which relate to mosquito reproduction and development.
The following procedures represent an optimized methodology that improves the efficiency of various steps in the protocol without loss of target-specific hybridization signals. Guidelines for RNA probe preparation, dissection of soft tissues and the general procedure for fixation and hybridization are described in Part A, while steps specific for the collection, fixation, pre-hybridization and hybridization of mosquito embryos are detailed in Part B.
Protocol
A. Hybridization in situ for Soft Tissues: Mosquito Salivary Glands and Ovaries
Recipes for solutions and buffers required for the following procedures are outlined in Table 1.
1. RNA Probe Preparation and Quality Analysis
- Design primers to PCR amplify the transcript target of interest. It is recommended that the amplicon is ≥600 nucleotides in length and more importantly that the amplicon sequence is unique to the target transcript. Sequences that are not-unique are avoided, to eliminate the possibility of generating cross-reactive RNA probes.
- Clone the PCR product into the pCR4-TOPO vector, or similar cloning vector. The pCR4-TOPO cloning vector contains T7 and T3 RNA polymerase priming sites that enable the production of both sense and antisense run-off transcription products from a single clone.
- Sequence the cloned amplicon to verify sequence fidelity and orientation within the vector plasmid.
- Perform a restriction digest reaction with an enzyme suitable to produce an antisense in vitro transcription product. Plasmid linearization is made straightforward by selecting a restriction site located in the multiple cloning region of the pCR4-TOPO vector such as SpeI, PstI or NotI. Ensure that the selected restriction site is not present in the cloned fragment. The same clone can be used to generate a sense-strand RNA probe, which serves as a background control. Hybridization in situ with a sense probe should give little or no signal, when compared to that of the complementary antisense probe.
- Verify by gel electrophoresis that the pCR4-TOPO clone is completely linearized.
- Purify the linearized plasmid DNA by performing a phenol-chloroform extraction, followed by ethanol precipitation. When precipitating the plasmid DNA it is recommended to use a final concentration of 2 M ammonium acetate with 2.5 volumes of ethanol to reduce the carry-over of residual salt into the in vitro transcription reaction.
- Prepare the digoxigenin (DIG)-labeled single-stranded RNA probe by performing a run-off in vitro transcription reaction, using 1 μg of the linearized plasmid DNA as a template. Prepare the reaction with RNA polymerase, transcription buffer and DIG RNA labeling mix as described by manufacturer’s instructions.
- Purify the DIG-labeled, in vitro transcription products by ethanol precipitation and resuspend the pelleted RNA in DEPC-treated distilled water.
- Verify the quality and expected molecular weight of the purified DIG-labeled RNA probe by formaldehyde gel electrophoresis. The prepared RNA probe can be divided into aliquots and stored at -80 °C until use.
2. Dissection of Mosquito Salivary Glands and Ovaries
- Dissect mosquito salivary glands using a probe and forceps as described previously for Ae. aegypti.1,2 For An. gambiae, salivary glands can be dissected as described for Ae. aegypti, or alternatively as described in the MR4 Methods for Anopheles Research manual.3
- Dissect mosquito ovaries using forceps as described previously for Ae. aegypti.4 Briefly, use one pair of forceps to grasp the thorax, while another pair of forceps pulls off the penultimate abdominal segment to release the ovaries.
- Collect dissected salivary glands or ovaries in Ambion RNAse-free 1.5 ml microfuge tubes containing 50 μl of PBS.
- Maintain samples on ice until fixation.
3. Fixation
- Fix dissected tissues in freshly-prepared soft tissue fixation solution (Table 1) at room temperature with nutation for 30 min to 1 hr. The fixation solution is prepared fresh to prevent oxidation of the formaldehyde fixative. Fixation for 1-1.5 hr is recommended for salivary glands. A minimum volume of 1 ml is recommended for fixing tissues in a 1.5 ml microfuge tube.
- Allow the tissues to settle to the bottom of the microfuge tube. This is an important step, which follows all changes of solutions in the protocol and will minimize significant sample loss.
- Decant the fixation solution by careful pipetting, leaving 50-100 μl of volume in the microfuge tube and then wash 3×, 5 min each with PBT.
- Optional equilibration steps for tissue storage. Stepwise rinse out PBT with either ethanol or methanol. Perform step-wise washes with PBT/alcohol (3:1, 1:1 and 1:3, 5 min each). Store in 100% (200 proof) alcohol at -20 °C. Tissues can be stored for several months without degradation of tissue ultrastructure. Before performing subsequent steps of the protocol, tissues must be returned to PBT by stepwise equilibration with alcohol/PBT (3:1, 1:1, 1:3, 5 min each).
- Perform PBT washes 3×, 5 min each.
- Perform protein digestion with 0.01 mg/ml Proteinase K/PBT, 5 min at room temperature. Alternatively, if both immunolocalization of protein targets and hybridization in situ of RNA are desired, ovaries can be incubated with 80% acetone/H2O at -20 °C for 10 min instead of treating with Proteinase K.
Proteinase K treatment is omitted for preparation of salivary glands. Proceed directly to step 3.10. - Immediately decant the digestion solution by careful pipetting. Wash 2×, 5 min each with ice-cold PBT to stop digestion.
- Perform additional PBT washes 2×, 5 min each at room temperature.
- Post-fix in soft tissue fixation solution at room temperature with nutation for 30 min. Post-fixation steps are not performed for salivary glands.
- Perform PBT washes 5×, 5 min each.
4. Hybridization
- Equilibrate tissues into hybridization solution (Hyb) by incubating at room temperature with 1 ml of Hyb/PBT (1:1) for 30 min with nutation.
- Decant as much Hyb/PBT as possible. Add 1 ml of Hyb into each sample tube.
- Wrap the sample tube in sealed-air protective packaging (bubble wrap) and place inside a hybridization bottle. Seal the bottle with a hybridization bottle cap.
- Pre-hybridization and hybridization steps can be performed easily in a hybridization oven. Perform pre-hybridization at 55 °C for 30 min with rotation.
- Allow sufficient time for the tissues to settle to the bottom of the microfuge tube, as the Hyb is more dense than PBT. Carefully decant the pre-hybridization solution. Tissues may become translucent and difficult to visualize. Sample loss can be reduced by leaving 50-100 μl of solution volume in each sample tube.
- Add 50 μl volume of Hyb into the sample tube and maintain in a heat block at 55 °C.
- Denature the RNA probe at 85 °C for 5-10 min. Immediately, place the denatured probe on ice for 5 min.
- Pipette the RNA probe into the sample tube. The RNA probe will float to the surface of the Hyb. Mix by flicking the tube gently.
- Perform hybridization in a total volume of 50-100 μl of Hyb per sample. Incubate the samples in a fixed microfuge rack inside a hybridization oven, or in a floating microfuge tube rack in a water bath, at 55 °C for 16-24 hr.
5. RNAse A Treatment
- Remove the Hyb containing the probe. It is important to maintain the samples at 55 °C during the wash steps to reduce non-specific binding of residual unbound probe. If several samples are hybridized, it is recommended to keep sample tubes in a heat block set at 55 °C to maintain the hybridization temperature throughout the washing steps.
- Perform a quick wash with Hyb by pipetting 1 ml of solution into the microfuge tube and inverting the sample tube 5-6 times.
All subsequent quick washes in the protocol are performed similarly by adding the required solution and inverting the sample tube several times. - Perform two additional washes with Hyb at 55 °C, 30 min each with rotation in the hybridization oven.
- Equilibrate into PBT by incubating at room temperature with Hyb/PBT (1:1) for 15 min with nutation.
- Perform PBT washes 5×, 5 min each.
- Prepare fresh RNAse A buffer. Perform RNAse A treatment by incubating in 20 μg/ml RNAse A/PBT at 37 °C, for 30 min. RNAse A cleaves single stranded RNA and will result in the degradation of unhybridized RNA probe.
- Decant RNAse A buffer and perform a quick wash with PBT.
- Perform PBT washes 3×, 5 min each.
6. Antibody Incubation
- Prepare fresh blocking solution (5% chicken serum/1% Western Blocking Reagent/PBT) and block samples by incubating in 1 ml of solution for 30 min at room temperature with nutation.
- Add 200 μl of a 1:1000 dilution of anti-DIG-alkaline phosphatase (AP)-conjugated antibody in blocking solution. Incubate at 4 °C, overnight with nutation.
7. Alkaline Phosphatase Staining
- Remove the antibody and blocking solution.
- Perform a quick wash with PBT.
- Perform additional PBT washes 5×, 10 min each. Alternatively the samples can be washed 3x, 10 min each followed by an extended wash at 4 °C, overnight with nutation.
- Prepare fresh AP staining buffer.
- Perform quick wash with AP staining buffer.
- Perform additional washes with AP staining buffer 3×, 5 min each with nutation.
- Prepare, according to manufacturer’s instructions, fresh AP staining solution containing the AP substrate NBT/BCIP.
- Incubate each sample with 500 μl of AP staining solution at room temperature with nutation, in the dark. The samples can be incubated by covering the tubes with an opaque container or aluminum foil. Reaction progression should be monitored every 1-2 minutes for the formation of a purple precipitate. Depending upon the probe concentration, concentration of target RNA and presence of non-specific targets, the staining may proceed for several minutes to several hours.
- Remove as much of the staining solution as possible. Unsatisfactory decantation results in the formation of a white flocculent precipitate during subsequent wash steps.
- The staining is stopped completely by performing two quick washes with PBT.
- Perform additional PBT washes 3×, 5 min each.
8. Glycerol Mounting
- Using a large-bore 200 μl pipette tip, transfer samples from microfuge tubes into individual round-bottom wells of a Pyrex spot plate.
- Carefully remove from the sample as much of the PBT as possible.
- Pipette on top of the sample 70% glycerol/H2O. Allow glycerol to permeate the sample tissue at 4 °C, overnight. Glycerol treatment prevents desiccation of tissues during slide mounting.
- Prepare microscope slides by wiping the slide surface clean with Kimwipes. Apply on to the slide two pieces of invisible tape parallel to each other so that they form a narrow channel (Figure 2).
- Using an artistic brush, pick up samples one-by-one and place them along the length of the channel, positioning each sample with the same orientation, with respect to anterior/posterior and dorsal/ventral alignments.
- After positioning each sample, use a Kimwipe swab to absorb excess glycerol solution surrounding the tissues (Figure 3). Removal of the excess glycerol is important to prevent the formation of bubbles, when sealing the mounted samples onto the slide under a cover slip.
- Center and place carefully a cover slip over the channel containing the aligned samples. Affix the cover slip semipermanently onto the slide by dabbing the corners of the cover slip with transparent nail polish.
- Using a 200 μl pipettor, dispense slowly into one end of the channel enough 70% glycerol to fill by capillary action the entire space of the channel and the area under the cover slip.
- Permanently seal the mounted samples by applying nail polish along all four sides of the cover slip. Allow the nail polish to dry sufficiently before photographing of the hybridized whole-mount tissues. It is recommended to photograph images of the samples immediately after mounting.
B. Hybridization in situ for Mosquito Embryos
Solutions and buffers required for hybridization in situ for mosquito embryos are described (Table 1). Fixation and hybridization procedures presented here have been modified from those first reported for Anopheles gambiae,5,6 Aedes aegypti7 and Culex quinquefasciatus.8
1. Embryo Collection
- Embryos are collected from 300 mated, female adult mosquitoes (allowed to mate with males for 48 h, 3 days post-emergence) 72 hr after blood feeding. The mosquitoes are maintained on 3 M sucrose solution, in order to prevent oviposition prior to the intended collection period.
- Prepare embryo collection containers by lining a 16 oz. paper cup with Saatilene Hitech mesh so that the inside surface of the cup is covered completely. The mesh can be affixed by using a single staple. Using this mesh greatly reduces fibrous contaminants that are present if Whatman filter paper, or paper towels are used.
- Fill the container half-full with distilled water.
- Embryos are collected for 1 hr by placing the collection container in the cage, which is subsequently covered by a dark cloth to stimulate oviposition.
- Collected embryos are allowed to mature to desired developmental stages outside of the cage in standard rearing conditions (85% relative humidity/ 26 °C). Maturation of embryos to various stages can be correlated to time (hours post-oviposition) following the approximate developmental time course of embryonic events described (Table 2). This table summarizes major events in embryonic development for Aedes aegypti, reported by Raminani and Cupp,9,10 Anopheles gambiae described first by Ivanova-Kazas11 and further elaborated by Goltsev and colleagues,12,13 and Culex quinquefasciatus reported by Davis.14 The time courses described are intended to serve as initial estimates, rather than absolute time points, for the collection of embryos at particular stages of development.
2. Dechorionation, Fixation, and Endochorion Disruption
- Transfer embryos from the collection container into a Saatilene mesh dechorionation catch tube (Figure 3).
For Aedes embryos, place the catch tube into a beaker that is filled with enough distilled water so that the volume of the catch tube is one-half full. An artistic brush can be used to dislodge the embryos from the Saatilene mesh and transfer them into the water-filled catch tube.
For Anopheles embryos, detach the Saatilene mesh from the collection container and fold the mesh carefully to form a funnel. Holding the mesh funnel with one hand, use the other hand to dispense distilled water through a polyethylene squirt bottle along the sides of the funnel to wash the embryos into the catch tube. This method can be applied to Anopheles embryos because they lack the adhesive substance present on the surface of Aedes embryos, which enables eggs to stick to oviposition substrates. - Prepare 40 ml of dechorionation solution (1 volume 5.25% sodium hypochlorite: 3 volumes of distilled water). Pour the solution into a 100 mm Petri dish.
- Prepare a 100 ml beaker containing 50 ml of distilled water and set aside. The dechorionation step is time-sensitive and immediately following sodium hypochlorite treatment the embryos will be immersed in this water, to dilute the dechorionation solution.
- Dechorionate the embryos in the mesh catch tube by placing the tube in the Petri dish filled with dechorionation solution. Use a disposable polyethylene transfer pipette or Pasteur pipette to wash the embryos, while swirling the catch vial so that the embryos remain submerged and agitated in the dechorionation solution.
A maximum of 35 sec is required for dechorionation of Aedes embryos, while 75 sec is sufficient for Anopheles and Culex embryos (Table 3). Dechorionation is one of the most sensitive steps of the protocol and over-extension of the sodium hypochlorite treatment, specifically for Aedes embryos will prevent proper cracking of the chorion. - Immediately submerge the catch vial in the beaker containing water.
- Wash off the remaining dechorionation solution from the embryos by removing the catch tube from the water-filled beaker and rinsing off the inner and outer walls of the catch tube with distilled water from a squirt bottle or faucet fitted with tubing.
- Dechorionation can be verified by visualizing the embryos with a dissecting microscope at low magnification. Dechorionated Aedes embryos lack the netting-like exochorion and only the smooth, polished surface of the black endochorion is apparent For dechorionated Anopheles embryos, the exochorionic floats and ridge structures are absent (Figure 4).
- Using an artistic brush, transfer the dechorionated embryos into a scintillation vial containing 5 ml of distilled water (Figure 5-1). If multiple samples are being prepared, maintain the catch tubes in water to prevent desiccation of the embryos.
- Let the embryos settle to the base of the scintillation vial and remove as much water from the vial as possible.
- Add 5 ml of heptane into the scintillation vial (Figure 5-2). Remove any residual water that remains in the scintillation vial.
- Add into the scintillation vial 5 ml of freshly-prepared embryo fixation solution (Table 1).
- Fix the embryos for 30 min with inversion so that the organic and aqueous phases mix thoroughly, but not vigorously.
- Remove the fixation solution using a glass Pasteur pipette, being careful not to disturb the interphase, which contains the embryos (Figure 5-3).
- Wash off the residual fixation solution by filling the scintillation vial with as much distilled water as possible. Invert the vial 5 times and then remove all of the distilled water.
- Fill the scintillation vial with 20 ml of distilled water and mix by inverting the vial for 30 min.
- Remove all of the water from the scintillation vial.
- Add enough boiling distilled water into the scintillation vial so that the inorganic heptane phase reaches the top of the vial.
- Incubate for 30 sec and then remove all of the boiling water.
- Add ice cold distilled water until the inorganic phase reaches the top of the vial. Incubate the vial in ice for 10 min.
- Remove first the aqueous and then organic phases. The heptane will appear opaque at this step (Figure 5-4).
- Add 5 ml of new heptane into the vial. Remove all remaining water from the vial.
- Add 5 ml of methanol into the vial. Swirl the vial 1-2 times energetically, without shaking (Figure 6). If the inorganic heptane and organic methanol phases are shaken vigorously the embryos will be destroyed during endochorion rupture. It is noteworthy to mention that we have observed variation in the efficiency of endochorion disruption for different strains of Aedes aegypti embryos.
- Incubate at room temperature for 15-20 min. During this stage, the phases will become turbid due to the release of yolk components (Figure 5-5).
- Remove both organic and inorganic phases and wash with methanol 3 times to remove all residual heptane.
- Embryos can be stored in methanol at -20 °C for up to several months.
3. Peeling
- Place a piece of double-sided toupee tape in the center of a 3 cm Petri dish. Toupee tape is used because the adhesive is stable in the presence of methanol and ethanol.
- Using a large bore (approximately 3 mm diameter) 200 μl pipette tip, transfer embryos from methanol onto the double-sided tape.
- Allow 1-2 min for the embryos to settle and adhere to the tape surface.
- Decant excess methanol and allow the surface to dry slightly for 1-2 min.
- Add 4 ml of 95% (190 proof) ethanol into the Petri dish.
- Pipette 600 μl of distilled water into the ethanol and swirl to mix. The tape will become opaque in color. Addition of distilled water will cause the tape to become tacky and immobilize the embryos during peeling. This step can be omitted if desired.
- Use a 27.5 gauge needle attached to a 1 ml syringe barrel to peel off the cracked endochorion (Figure 7). After releasing the white embryo from the black endochorion remnants, transfer the embryos from the tape to the ethanol reservoir of the Petri dish using a fine-tip artistic brush. Using a 3-mm bore pipette tip, transfer the embryos into an Ambion 1.5 ml microfuge tube containing 500 μl of 200 proof ethanol.
The peeled embryos should not remain on the tape for an extended period of time because they may adhere permanently to the tape. - Perform 2-3 quick washes with 100% (200 proof) ethanol and store the embryos in punctilious ethanol at -20 °C. Peeled embryos can be stored at -20 °C for a few months without affecting morphology or the subsequent hybridization signal.
4. Clarification of Yolk
- Perform a quick wash with 200 proof ethanol. Punctilious ethanol should be used for all subsequent wash steps and for preparation of the P-xylene yolk clarification solution.
- Perform one ethanol wash, 5 min with nutation.
- Incubate in P-xylene/ethanol (9:1) at room temperature for 1-1.5 h with nutation. The xylene step permits clarification of the yolk mass to enhance the signal to noise ratio.
- Remove the xylene-ethanol solution and perform two quick washes with ethanol.
- Perform one ethanol wash, 5 min with nutation.
- Perform two quick washes with methanol, followed by one methanol wash, 5 min with nutation.
- Equilibrate into 4% formaldehyde fixation solution by washing once with methanol/ formaldehyde fixation solution (1:1).
5. Fixation and Hybridization in situ
Fixation and hybridization in situ procedures are identical to those described in protocol Section A.
C. Representative Results
The hybridization in situ protocol described here, results in colored staining patterns that indicate presence and localization of the targeted mRNA. It is important to emphasize that relative levels of transcript abundance cannot be determined by hybridization in situ. Hybridization results are dependent upon the specific mRNA probe used for the hybridization procedure, the abundance of the target mRNA in the hybridized tissue, as well as probe concentration and hybridization temperature. Comparison of tissues hybridized with antisense and corresponding sense mRNA probes make possible the accurate interpretation of staining patterns.
Hybridization in situ of whole-mount salivary glands of female Aedes aegypti with mRNA probes that target amylase1 (AAEL006719), D7s2 (AAEL006423), and D7L2 (AAEL006424) indicate accumulation of these transcripts in proximal-lateral, distal-lateral, and distal-lateral/medial lobes, respectively (Figure 8).1 Whole-mount ovaries of three mosquito species were hybridized with mRNA probes that specifically target the respective orthologous transcripts of oskar (Figure 9).7,8 Hybridization in situ of whole-mount embryos of Ae. aegypti, An. gambiae and Cx. quinquefasciatus was performed using antisense RNA probes targeting the respective mosquito oskar orthologous transcripts (Figure 10).7,8
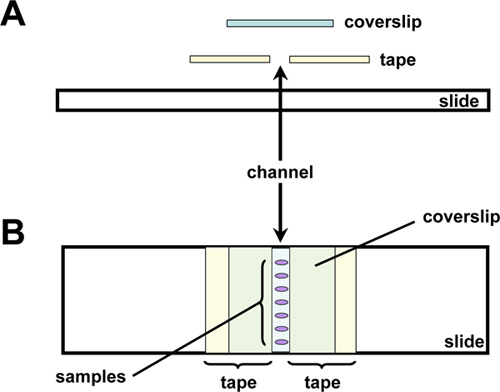
Figure 1. Schematic diagram of post-hybridization mounting set-up.
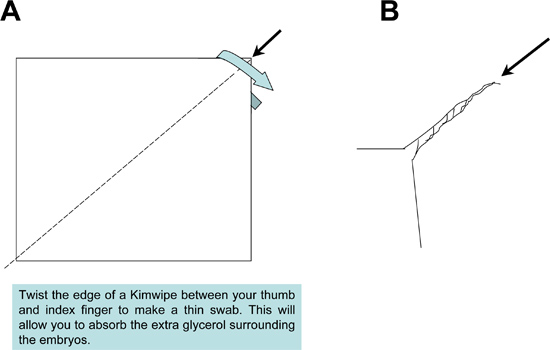
Figure 2. Preparation of Kimwipe mop used during sample mounting. A Kimwipe tissue is twisted tightly to produce a fine-tipped mop to absorb excess mounting medium from the samples and slide.
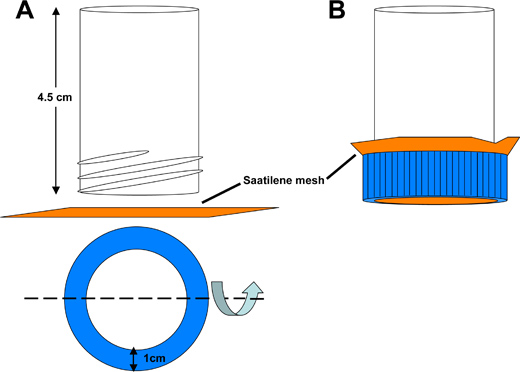
Figure 3. Schematic of Saatilene mesh dechorionation catch tube. A) The bottom of a 50 ml polystyrene conical tube is cut off to yield a hollow tube 4.5 cm in length, open at both ends. A circular opening is cut out of the conical tube lid to permit liquid to be washed through a 6.5 cm2 square piece of Saatilene mesh. The 330 threads per inch, 34 micron diameter thread mesh screen retains the mosquito embryos. B) Assembled catch tube.
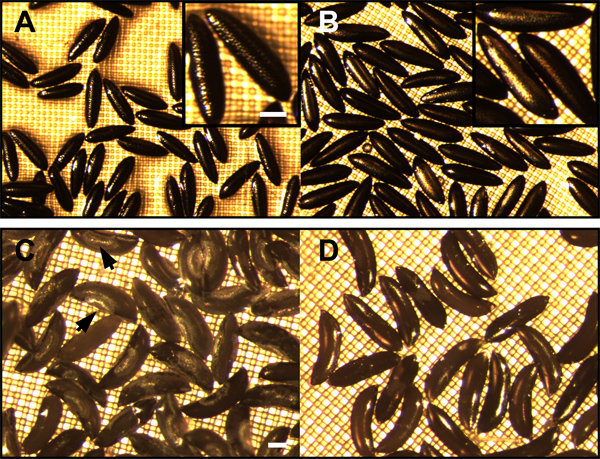
Figure 4. Aedes aegypti and Anopheles gambiae eggs, before and after dechorionation. A) Aedes aegypti eggs prior to dechorionation. The mesh-like exochorion lies above the black endochorion and gives the egg a textured appearance (inset enlargement). Following removal of the exochorion, only the smooth and polished endochorion remains (B and inset enlargement). C) Eggs of Anopheles gambiae prior to dechorionation. Exochorion structures such as floats are visible (arrows). D) The smooth and polished surface of the endochorion is visible after dechorionation. Bar = 100 μm.
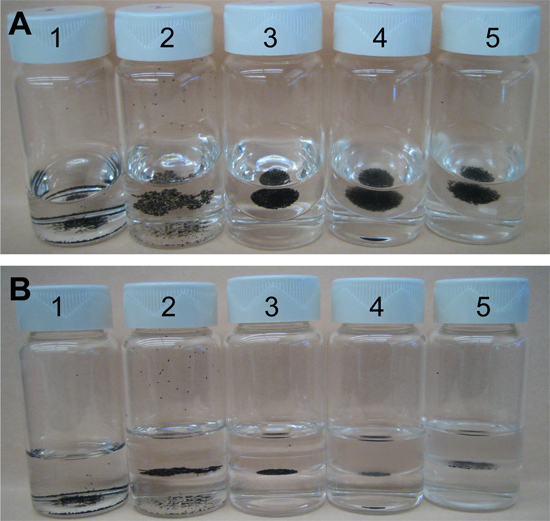
Figure 5. A series of sequential steps for fixation and endochorion disruption of Ae. aegypti eggs. A) Slanted-frontal view and B) lateral-view of scintillation vials containing Ae. aegypti eggs during sequential steps of fixation and endochorion disruption. 1) Embryos in distilled water. 2) Embryos float in the interphase between the upper heptane phase and the lower aqueous phase. 3) Following fixation the embryos pack together in a round mass. Embryos remain in the interphase between heptane and fixative solution phases. 4) After treatment with boiling water and incubation in ice, the heptane phase becomes slightly opaque. 5) Embryos with disrupted endochorions are shown in the interphase between an opaque heptane phase and transparent methanol phase.
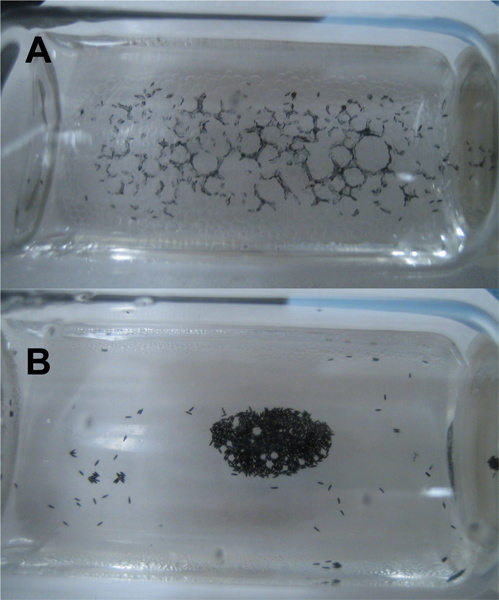
Figure 6. Endochorion disruption of fixed Ae. aegypti eggs. A) Immediately after energetic swirling of heptane and methanol phases, the formation of bubbles and the disruption of the endochorion can be visualized. B) Following five minutes of incubation, at room temperature.
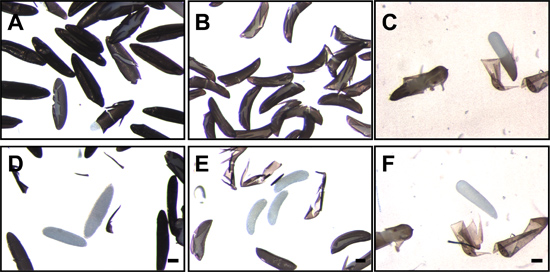
Figure 7. Mosquito eggs following disruption and removal of the endochorion. Eggs of Ae. aegypti (A), An. gambiae (B) and Cx. quinquefasciatus (C) following fixation and disruption of the endochorion. White, translucent embryos can be seen within the cracked endochorion. After removal of the endochorion, the translucent embryos of Ae. aegypti (D), An. gambiae (E) and Cx. quinquefasciatus (F) are clearly visible. Bar = 100 μm.
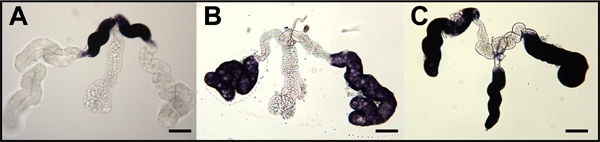
Figure 8. Hybridizations in situ for three genes expressed in different lobes of whole-mount, female Ae. aegypti salivary glands. Staining is indicative of localization and accumulation of amylase1 (AAEL006719) (A), D7s2 (AAEL006423) (B) and D7L2 (AAEL006424) (B). Bar = 100 μm.
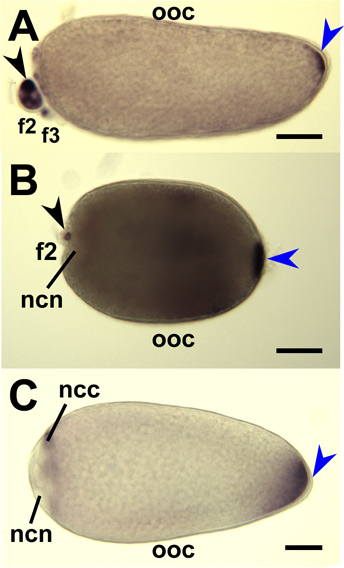
Figure 9. Hybridization in situ for mosquito oskar antisense RNA probes to whole-mount mosquito oocytes and nurse cells. Stage IV oocytes (ooc) dissected from ovaries of An. gambiae (A), Ae. aegypti (B) and Cx. quinquefasciatus (C) and hybridized with RNA probes targeting respective mosquito oskar mRNAs. Primary follicles are oriented with anterior on the left. Staining at the posterior pole (blue arrowhead) indicate accumulated oskar mRNAs. Secondary (f2) and tertiary (f3) follicles are shown and staining (black arrowhead) indicate accumulation of oskar mRNA in the nurse cell cytoplasm (ncc). Staining is excluded from the nurse cell nuclei (ncn). Bar = 50 μm.
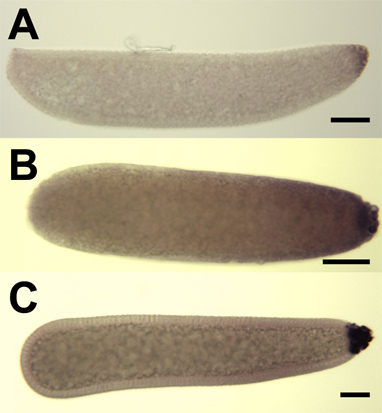
Figure 10. Hybridization in situ for mosquito oskar antisense RNA probes to whole-mount mosquito embryos. Embryos are oriented with anterior on the left. Cellular blastoderm stage embryos of An. gambiae (A) and Cx. quinquefasciatus (C) are hybridized with respective mosquito species-specific oskar RNA probes. B) A syncytial blastoderm stage Ae. aegypti embryo hybridized with RNA probes targeting Ae. aegypti oskar transcript. Staining is evident in the posterior pole cells of all embryos, indicating localization and accumulation of mosquito oskar mRNA in these cells. Bar = 50 μm.

Table 1. Solutions and buffers for formaldehyde gel electrophoresis, fixation and hybridization in situ.
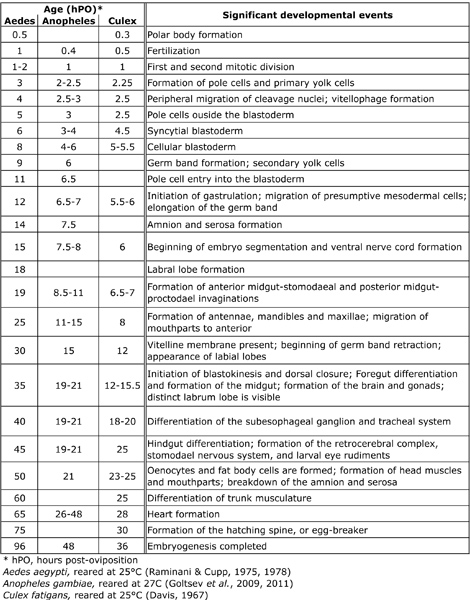
Table 2. Developmental events and morphological observations corresponding to consecutive stages during mosquito embryogenesis.
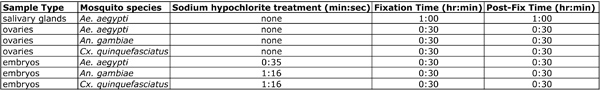
Table 3. Summary of key differences in pre-hybridization steps for various tissue types.
Discussion
The hybridization in situ and colorimetric staining protocol presented here for whole-mount mosquito tissues and embryos is a useful technique for the localization of transcripts within specific organs and cell types. These procedures are an improvement over our previously reported methods, both in streamlining extensive wash steps and providing additional technical details and reagent sources.
In our experience, colorimetric detection of hybridization signals is superior in sensitivity and clarity of hybridization signal compared to fluorescence-based detection schemes. Moreover colorimetric detection circumvents issues associated with signal discrimination in embryos, which are inherently auto-fluorescent. Limitations in detecting hybridization signals occur, when low-abundance transcripts are targeted and background staining is evident. Increasing the hybridization temperature to 65 °C has been found to reduce background hybridization signals, but is not a suggested substitute for designing unique target-specific RNA probes.
This protocol has been used to perform hybridization in situ of whole-mount salivary glands of Aedes aegypti, and ovaries and embryos of Anopheles gambiae, Anopheles stephensi, Ae. aegypti and Culex quinquefasciatus. This method also is applicable to other mosquito tissues, and presumably those of other insects. Additionally, we compiled for the first time comparative guidelines for the staging of embryonic development in three vector mosquitoes, Ae. aegypti, An. gambiae and Cx. fatigans. Observations have been reported for specific strains of these three species, under discreet rearing conditions. It is important to note that the developmental time course can vary for different strains of mosquito species and under different rearing conditions. Hybridizations in situ supplement on-going efforts to analyze the transcriptomes of mosquitoes and other arthropods, and may provide a better picture of the regulation of gene expression in these organisms.
Declarações
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Marika Walters for advice in developing hybridization in situ methods for soft tissues and Yury Goltsev for discussion of protocols for hybridization in situ of Anopheles gambiae embryos, which were subsequently adapted and modified to develop the protocol described here for hybridization in situ of Aedes and Culex embryos. We also acknowledge helpful recommendations given by Adam Paré and David Kosman. We thank Osvaldo Marinotti for scientific discussion and editing the protocol text.
Materials
| Name of the reagent | Company | Catalogue number | Comments (optional) |
| 0.5 M EDTA | Ambion | AM9261 | |
| 1M Tris-HCl | Ambion | AM9855G | pH8.0 |
| 10X PBS | Ambion | AM9625 | |
| 20X SSC | Ambion | AM9763 | |
| 1.5 ml microfuge tubes | Ambion | AM12400 | Less opaque than standard tubes; aids in visualizing samples |
| Deionized formamide | Ambion | AM9342 | Storage at 4 °C |
| DEPC water | Ambion | AM9932 | |
| Proteinase K | Ambion | AM2546 | |
| 5.25% Sodium hypochlorite | Austin’s | A-1 Brand | |
| T3 RNA Polymerase-Plus | Ambion | AM2733 | Storage at -20 °C |
| T7 RNA Polymerase | Ambion | AM2082 | Storage at -20 °C |
| 95% Ethanol | Fisher Scientific | AC61511-0040 | |
| Fisherbrand disposable polyethylene transfer pipettes | Fisher Scientific | 13-711-7M | |
| 37% Formaldehyde | Fisher Scientific | F79-500 | |
| HPLC-grade methanol | Fisher Scientific | A452-1 | |
| Magnesium chloride | Fisher Scientific | M87-500 | |
| Microscope cover glass | Fisher Scientific | 12-542A | 18 x18 mm |
| N-Heptane | Fisher Scientific | H350-1 | |
| P-xylene | Fisher Scientific | O5082-500 | |
| Pyrex 9-well Spot Plate | Fisher Scientific | 13-748B | 100×85 mm |
| Sodium chloride | Fisher Scientific | AC32730-0025 | |
| Sodium hydroxide | Fisher Scientific | SS255-1 | |
| Superfrost/Plus microscope slides | Fisher Scientific | 12-550-15 | 25x75x1.0 mm |
| Davlyn Red Clear-liner Toupee tape | Hair Direct | RED-75R12 | Poly/Skin base material 0.75 in x 12 yd tape roll |
| TOPOTA Cloning Kit for Sequening with One Shot Top10 chemically-competent E. coli | Invitrogen | K457501 K457540 | 20 reactions 40 reactions |
| Sonicated salmon sperm DNA | Invitrogen | 15632-011 | Storage at -20 °C |
| Anti-digoxigenin-AP Fab fragments | Roche Applied Science | 1093274 | Storage at 4 °C |
| DIG RNA labeling mix | Roche Applied Science | 1277073 | Storage at -20 °C |
| NBT/BCIP stock solution | Roche Applied Science | 1681451 | Storage at 4 °C |
| Western Blocking Reagent | Roche Applied Science | 11921673001 | Storage at 4 °C |
| Saatilene Hitech polyester mesh (330.130) | Saati Print | 330.34 UO PW | 330 threads/inch, 34 micron thread diameter, orange color |
| Glycerol | Sigma | G6279-1 | 70% in PBT |
| Heparin sodium salt | Sigma | H3393 | |
| Tween 20 | Sigma | P1379-500 | |
| 37% Formaldehyde | Ted Pella | 18508 | 10 ml aliquots in amber ampoules |
| 16 oz. Solo Paper containers with lids | The Paper Company | SOLOKH16AJ8000 | |
| Borosilicate glass scintillation vial with unattached screw cap | VWR International | 66022-128 | 20 ml case of 500 |
| Sealed Air Bubble wrap celluar cushioning material | VWR International | 500018-081 | 10 foot/roll 0.188 inches thick |
| Chicken serum | Whole blood was collected either from the wing vein or by cardiac puncture from a juvenile chicken. Blood was incubated at 37 °C for 1 h until coagulated and then placed on ice for 30 min. The serum was collected and centrifuged at 3000 x g for 10 min. The resulting supernatant (clarified serum) was collected and stored at -20 °C until use. |
Table 4. Table of specific reagents and equipment for hybridization in situ.
Referências
- Juhn, J. Spatial mapping of gene expression in the salivary glands of the dengue vector mosquito, Aedes aegypti. Parasit. Vectors. 4, 1 (2011).
- Coleman, J., Juhn, J., James, A. A. Dissection of midgut and salivary glands from Ae. aegypti mosquitoes. J. Vis. Exp. (5), e228 (2007).
- Benedict, M. Q. Dissecting Plasmodium-infected Mosquitoes: Salivary Glands, Chapter. 5.4.2. Methods in Anopheles Research. , (2007).
- Sappington, T. W., Brown, M. R., Raikhel, A. S. Culture and analysis of insect ovaries, Chapter. 42.3. The Molecular Biology of Insect Disease Vectors: A methods manual. , (1997).
- Goltsev, Y., Hsiong, W., Lanzaro, G., Levine, M. Different combinations of gap repressors for common stripes in Anopheles and Drosophila embryos. Dev. Biol. 275, 435-446 (2004).
- Benedict, M. Q. Anopheles Embryo Fixation, Chapter. 3.7. Methods in Anopheles Research. Malaria Research and Reference Reagent Resource Center (MR4). , (2007).
- Juhn, J., James, A. A. oskar gene expression in the vector mosquitoes, Anopheles gambiae and Aedes aegypti. Insect Mol. Biol. 15, 363-372 (2006).
- Juhn, J., Marinotti, O., Calvo, E., James, A. A. Gene structure and expression of nanos (nos) and oskar (osk) orthologues of the vector mosquito, Culex quinquefasciatus. Insect Mol. Biol. 17, 545-552 (2008).
- Raminani, L. N., Cupp, E. W. Early Embryology of Aedes aegypti (L.) (Diptera: Culicidae). Int. J. Insect Morphol. & Embryol. 4, 517-528 (1975).
- Raminani, L. N., Cupp, E. W. Embryology of Aedes aegypti (L.) (Diptera: Culicidae): Organogenesis. Int. J. Insect morphol. & Embryol. 7, 273-296 (1978).
- Ivanova-Kazas, O. M. Embryonic development of Anopheles maculipennis Mg. Izv. Akad. Nauk SSSR ser. Biol. 2, 140-170 (1949).
- Goltsev, Y. Developmental and evolutionary basis for drought tolerance of the Anopheles gambiae embryo. Dev. Biol. 330, 462-470 (2009).
- Papatsenko, D., Levine, M., Goltsev, Y. Clusters of temporal discordances reveal distinct embryonic patterning mechanisms in Drosophila and Anopheles. PLoS Biol. 9, e1000584 (2011).
- Davis, C. W. C. A comparative study of larval embryogenesis in the mosquito Culex fatigans Wiedemann (Diptera: Culicidae) and the sheep-fly Lucilla seriata Meigen (Diptera: Calliphoridae) I. Description of embryonic development. Aust. J. Zool. 15, 547-579 (1967).

