Using Flatbed Scanners to Collect High-resolution Time-lapsed Images of the Arabidopsis Root Gravitropic Response
Summary
This protocol describes a process for rapid collection of images of Arabidopsis seedlings responding to a gravity stimulus using commercially-available flatbed scanners. The method allows for inexpensive, high-volume capture of high-resolution images amenable for downstream analysis algorithms.
Abstract
Research efforts in biology increasingly require use of methodologies that enable high-volume collection of high-resolution data. A challenge laboratories can face is the development and attainment of these methods. Observation of phenotypes in a process of interest is a typical objective of research labs studying gene function and this is often achieved through image capture. A particular process that is amenable to observation using imaging approaches is the corrective growth of a seedling root that has been displaced from alignment with the gravity vector. Imaging platforms used to measure the root gravitropic response can be expensive, relatively low in throughput, and/or labor intensive. These issues have been addressed by developing a high-throughput image capture method using inexpensive, yet high-resolution, flatbed scanners. Using this method, images can be captured every few minutes at 4,800 dpi. The current setup enables collection of 216 individual responses per day. The image data collected is of ample quality for image analysis applications.
Introduction
Collection of high-resolution phenotypic data is useful in studies that aim to understand the interplay of genetics and environment in mediating organismal function1,2. Studies of this nature are also inherently large in scale, making it additionally necessary that methods employed for measuring phenotypes in this context be high in throughput3,4. In establishing methods for phenomics-scale research, tradeoffs between throughput and resolution come into play. Methods that are higher in throughput also tend to be lower in resolution, making it more difficult to detect small effects of genetics or environment5. Alternatively, methods that more carefully measure a desired phenotype also tend to be lower in throughput, making it difficult to survey genetic and environmental effects broadly. Additionally, manual methods for quantifying phenotypes, including visual inspection, can be subject to variation due to differences in human perception6.
Imaging technologies can provide a useful bridge between throughput and resolution in obtaining phenotypic observations7-9. In general, an image is relatively easy to capture, facilitating throughput, and when taken at sufficient resolution, subtle phenotypes can be detected1,2,7. Imaging technologies tend to be modifiable to fit a system or process of interest and are generally scalable10-12. Because of this, imaging technologies are ideal for the development of large-scale studies of organismal function.
The response of the primary root to a gravity stimulus is an intricate physiological process that occurs within a morphologically simple organ. The response involves activation of signaling pathways that propagate through the root organ and its progression is determined by environmental and genetic factors, including genetic factors influenced by the environment12-14. The response of the primary root to a gravity stimulus has been studied at least since Darwin, yet there is much to learn about how it works, particularly in the early signaling events and in the factors mediating response plasticity12,14,15. Gaining a detailed understanding of the dynamics of this response is important in finding ways to improve the ability of seedlings to successfully become established within a given environment16. In addition, the shape of the root makes it amenable for image processing applications8,12,17. Taken together, the root gravitropic response is an ideal system for the development of high-throughput imaging technology for the purpose of conducting genomics-level studies of organismal function.
In this report, a high-throughput, high-resolution method for image capture of the root gravitropic response using inexpensive, commercially-available flatbed scanners is presented. The overview of the protocol is shown in Figure 1. Seedlings planted on agar plates were positioned on vertically-oriented flatbed scanners fitted with custom Plexiglas plate holders. Images were collected every few minutes at 4,800 dpi and saved on a local drive or data server. Metadata associated with each image series is stored on a database and the stored images are processed. The approach uses the VueScan software for image capture. VueScan can be used to run over 2,100 different scanners on Windows, Mac, or Linux operating systems (see Materials Table). A scanner resolution of 4,800 dpi was used in this application to match the resolution achieved in previous studies using fixed CCD cameras1,8,12. The flexibility of the VueScan software along with the common interface it uses for any scanner it runs allows users to readily adopt virtually any scanner hardware of sufficient resolution to the protocol presented in this paper. Current throughput allows for the collection of 216 individual responses per day. The technology is adaptable and scalable for use at institutions ranging from high schools to research universities. Furthermore, the images collected are of sufficient quality for image analysis applications.
Protocol
1. Image Acquisition Protocol
Considerations:
This protocol is most efficiently performed with two people, although it is possible for one to work alone. The arrangement working best in this laboratory was for one person to prepare plates for scanning while another works on scanner setup, then both work together to place plates in scanners and start the scanning process.
It's also important to note that the scanners in this project are vertically oriented with the scanner lids resting on the back of the scanner. A custom support was made to hold dishes in this vertical position and was affixed to the flatbed surface with 3M Command Strips (Figure 2). The removable document cover that comes with the scanner used in this protocol (an Epson V700) was lined on one side with black felt. The document cover was positioned against the flatbed with a bungee cord to hold the plates in place and to provide image contrast (Figure 3).
Any scanner of sufficient resolution could be used for image capture. The Epson Perfection V700 was chosen because of its square profile (making it easy to position vertically), its high resolution, and the additional options to scan from the both the bed and lid and to use the infrared channel. These additional options were not used in this protocol.
Once the plates have been removed from the growth chamber, it is imperative that the protocol continue to the end.
Plate Preparation
Standard Petri dishes containing 10 ml of transparent medium and 9 seeds planted across the middle of each plate were used. Procedures for plate labeling, media preparation and planting can be found at: http://www.doane.edu/doane-phytomorph
- Retrieve the first agar plate and absorb collected condensation on the lid and rim of the lid of the agar plate with a Kimwipe.
- Apply Triton X-100 (a detergent) to lid with a Kimwipe — be generous.
(Note that Triton X-100 helps prevent the buildup of condensation on the lid as the plate is scanned. A generous application (enough to create a film on the lid surface) will help make sure that the lid stays transparent throughout the entire scanner run.) - Wrap the plate with micropore tape to secure the lid, and to allow for ventilation.
Scanner Setup and Image Collection
This protocol assumes that more than 1 scanner is being used, and provides instructions to start multiple scanners from a single computer.
- Create folders for storing images from each scanner. Each scanner will hold two plates, so keep this in mind when creating folders. One might choose to use metadata as components of the file name such as unique IDs for each plate, seedling ages, seed size, and IDs of stocks planted. An example of a folder name used in data collection containing these metadata is "1652-2-sm-9-92-17-1653-2-lg-88-79-161".
- Set outlet timers for designated collecting time (9 hr was used in this laboratory). Make sure to set extra time (an hour or so) for preparation.
(Note that scanners should be plugged into outlet timers in order to set the acquisition time. While the VueScan software allows a user to collect images repeatedly, it does not allow the user to indicate how many images to collect or how long to collect images for.) - Turn on the first scanner and wait approximately 10 sec for the scanner to go through its initial warm-ups.
- Open the VueScan program once. VueScan version 9.0.20 was used in this protocol (see Materials Table), though more recent versions can be used with little modification. Make sure the 'More' button has been pressed on the bottom panel of the user interface in order to display the menu options described below.
- Set the Auto repeat: drop-down box to none under the Input tab and under the Crop tab set Preview area: to Maximum (Figure 4). Press 'Preview'.
- Create a crop box that would capture the region of interest by using the mouse to click and drag across the region of interest on the preview image. Settings may be altered for the region of interest in the Crop tab. The typical settings used for the crop box were: x-offset 0.675; y-offset 1.924 in, though this was adjusted to capture the seedling area for each scanner. The crop box size used was 7.246 in wide by 1.1 in tall (Figure 5).
- To move the crop box, hold the shift key while dragging with the mouse. Make sure the crop box contains all the seedlings to be scanned plus any desired metadata that might be contained on a label (Figure 5).
- Under the Crop tab, set the Preview area: para Crop box and Press 'Preview'.
- Go to the Output tab and select the correct file for the scanner (Figure 5).
- Repeat steps 1.7-1.12 on all scanners for one computer. Choose the 'yes' option when asked whether to open more than one instance of VueScan.
- Go through each tab and verify the settings are correct. (Note that all specifications can be altered to fit the needs of an individual laboratory including image color, resolution, etc. However, the settings used in this protocol can be directly applied to the particular scanning hardware of a given lab due to the common interface of the VueScan software. Refer to the attached specifications list to view the parameters used in this project, using VueScan version 9.0.20).
- Under the Input tab choose Continuous in the Auto repeat: field, or choose a longer time interval between images if desired. The time interval is the length of time the scanner pauses after saving the last image and beginning collection of the next image. In Continuous mode, 3-4 min resolution can be obtained at 4,800 dpi.
- Repeat steps 1.14-1.15 for the rest of the scanners connected to a single computer.
- Place prepared plates in the correct scanners with seedlings oriented horizontally (do not gravistimulate).
- Temporarily place a black, felt background against the plates so they do not fall from Plexiglas template. Repeat for all scanners.
(Note: In this project, black pieces of felt were attached to the document covers provided with the equipment to prevent glare and to provide contrast against root tissue. The specific background color used will depend on the color of tissue being imaged). - Have one person turn the plates 90° (plates were turned counterclockwise in this protocol) and immediately replace the felt background.
- The other person should be standing at the computer so that they can immediately press the 'Scan' button.
- Secure the background to the scanner with a bungee cord (Figure 3). Have one person hold the background in place while another positions the bungee cord.
(Note: Immediately after gravistimulation (rotation of the plates by 90°) and placement of the felt background, 'Scan' should be pressed). - Repeat steps 1.17-1.21 for the rest of the scanners on a single computer.
- Repeat steps 1.6-1.22 for the next set of scanners if applicable.
- Do not leave the scanners until several images have been collected to make sure they are saving correctly.
- It is ideal to keep the scanners in an area that will be free of disturbances for the designated scan time. It is also prudent to consider the environmental conditions in the scanning area to ensure ideal phenotypic responses.
- When data collection is complete, press the green abort button on each VueScan window that coincides with each scanner.
- Close out of all programs on the computer.
- Restart the computer and shut off all the scanners before beginning another round of image collection.
Representative Results
Representative Images
This approach enables rapid production of high-resolution time series of Arabidopsis seedling growth. First and last images of a scanner run are shown in Figures 7A and 7B. Figures 7C and 7D show optimal results from half of a full scanner image. Some issues that can affect image quality are shown in Figures 7A and 7B. These issues include variation in germination, variation in seedling growth trajectory at the start of the run, and buildup of condensation during scanning. Condensation can largely be resolved by increasing the amount of Triton X-100 applied to the inside of the plate lid. Other factors that could inhibit accurate image collection are incorrect configuration of the crop box with respect to the plate position and positioning plates such that they are skewed with respect to the crop box.
Image Analysis Application: Image Compression
Once a time sequence of scanner images has been obtained, it must be securely stored in a network accessible manner to facilitate image analysis. The image files associated with an individual scanner run occupy a significant amount of hard drive space. A single TIFF file collected at 4,800 dpi is about 220 MB and a typical scanner run generates 200 image files. Therefore, about 44 GB of hard drive space is required per run. To reduce storage and network transmission costs associated with image analysis it is desirable to reduce the amount of space needed to store image data while at the same time minimizing data loss. Downstream analysis will involve identification of each seedling in subsequent image files associated with an experimental run. Therefore, segmenting out single seedlings from the scanner image can facilitate downstream analysis. Because segmentation of the seedling away from the rest of the image can also significantly reduce storage of unnecessary background pixels, this approach also leads to significant reduction in data size. Furthermore, if downstream analysis is focused on root tissue it may not be necessary to retain color information since the root pixels are relatively narrow in their color space. A computer image processing protocol and code to reduce data size by both segmenting out individual seedlings and converting images to grey scale has been developed. The approach results in a 60% reduction in storage space requirements.
The workflow used to achieve this data compression is described in the following steps:
- Start with a time series of scanner image files in a single folder.
- For each image, convert from an RGB to grey scale (Figure 8, top).
- Split the image into left and right sides.
- Extract each seedling from the image into its own file (Figure 8). This is done by applying a threshold to convert pixels to black or white and then calculating the total pixel intensity of each image row. The row with the highest intensity is identified and each pixel is classified as 'plant' or 'nonplant' based on the intensity of its neighbors. The center of each 'plant' within this row is found and from that point a crop box of a predetermined size is drawn (Figure 8, bottom).
- Create a separate folder for each side of the image (left and right) with separate subfolders for each seedling for storage of individual time series image files.
- Archive the resulting folders into a compressed ZIP file.
A code that accomplishes these steps has been developed using the programming language Python20. The algorithm allows for an approximately 60% reduction in data size and is successful in in identifying all individual seedlings in 90% of the scanner image files analyzed thus far. The codes are freely available for download under the GNU General Public License Version 3 (see Materials Table).
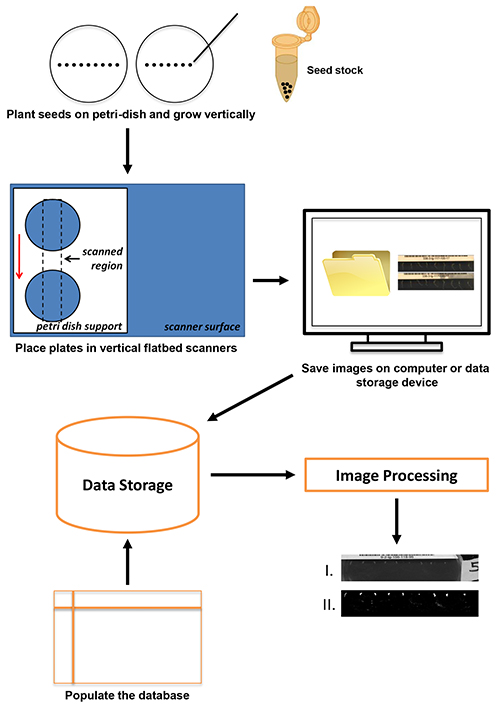
Figure 1. The scanning procedure begins with seed planting (up to nine Arabidopsis seeds per plate) and ends with data storage and image processing. Click here to view larger image.
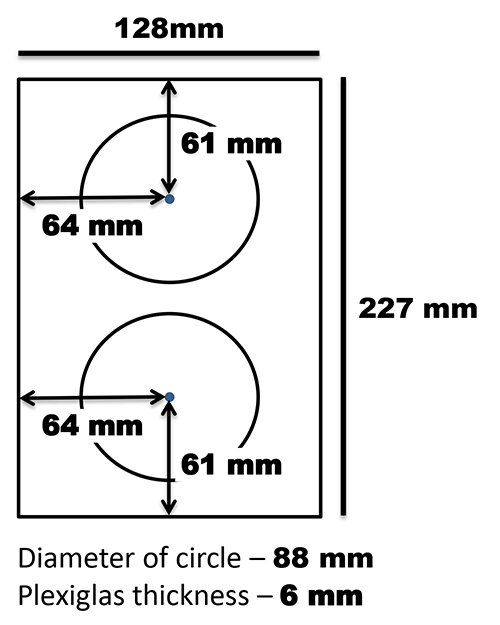
Figure 2. Template for construction of Petri dish support. Plexiglas was cut such that the width fit the flatbed (in this case 227 mm) and the length was 128 mm. Two circles with an 88 mm diameter were cut out of the remaining piece such that they were evenly distributed along the width and length of the support. The support was affixed to the flatbed with 3M Command Strips. Click here to view larger image.
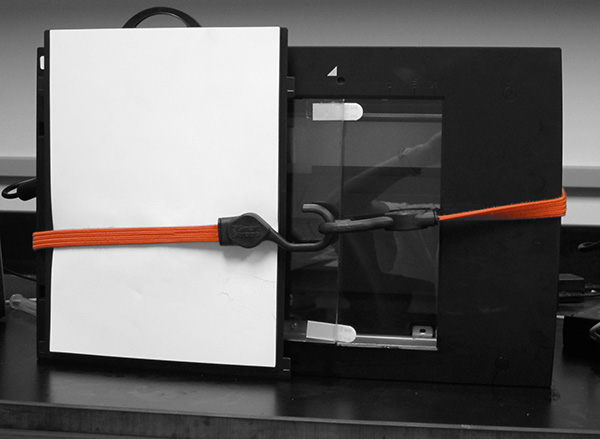
Figure 3. Scanner configuration after seedlings have been gravistimulated and the document cover positioned. This is the configuration of the scanner at step 1.21 of Scanner Setup and Image Collection. Click here to view larger image.
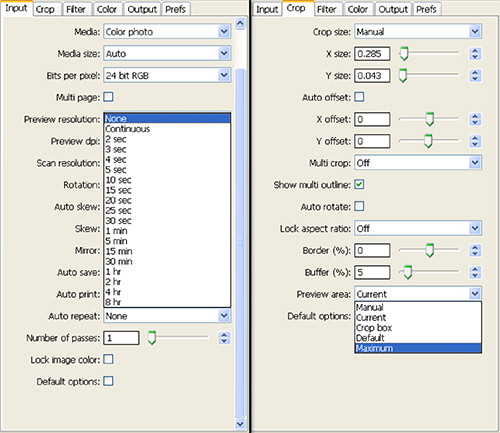
Figure 4. Screen shot of settings selected for step 1.8 of Scanner Setup and Image Collection. Click here to view larger image.
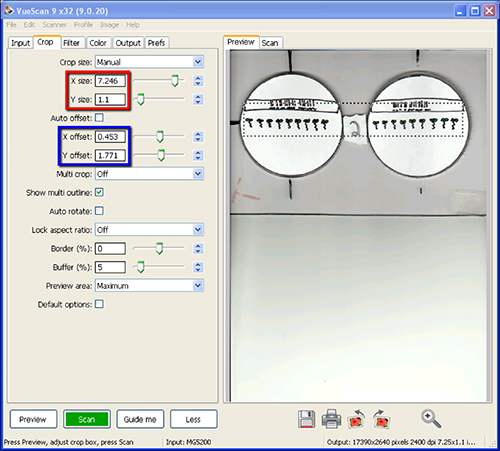
Figure 5. Screen shot of VueScan software during steps 1.9 and 1.10 of Scanner Setup and Image Collection. The red box highlights the crop size while the blue box highlights specific settings for x- and y-offset used in order to capture seedlings and label information. The region of the flatbed to be scanned is shown as a dotted line in the Preview area. Click here to view larger image.
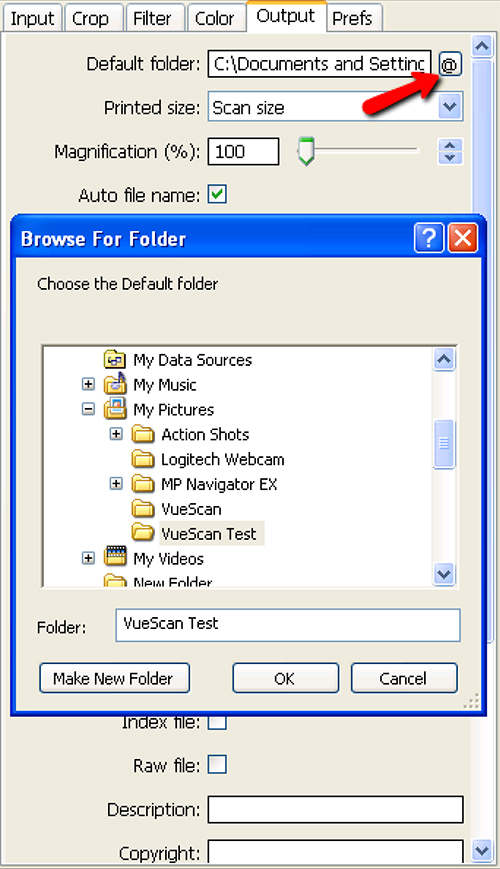
Figure 6. Selection of destination folder for step 1.12 of Scanner Setup and Image Collection. Pressing the @ button next to the Default Folder dialog box (red arrow) allows the user to select the appropriate destination folder. Click here to view larger image.
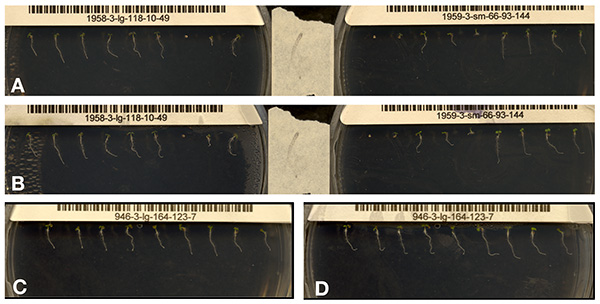
Figure 7 (A-D). The above images are examples of those collected using the method described in this paper. Panels A, B and C, D are the first and final images, respectively, from a single scan period. A, B show the full scanned area, while C, D are a cropped region of the scanned area, showing a single plate. Several inconsistencies can be observed. Panel A shows variation in germination and in growth trajectory. Panel B (the same seedlings as Image A; 9 hr later) shows that plates can accumulate condensation. Panels C and D are considered to be good results due to robust growth of seedlings and image quality throughout the run. Click here to view larger image.
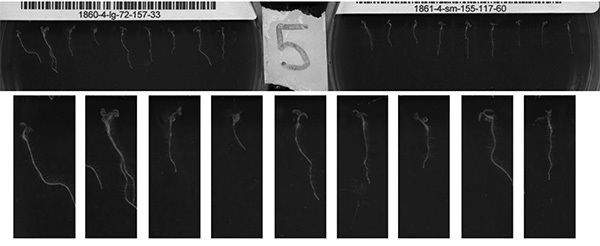
Figure 8. The image compression algorithm developed converts a scanner image to grey scale (top). The image is divided into right and left halves and image borders are removed (not shown). The positions of individual seedlings on each half are identified by finding the row with the largest total pixel intensity. Those positions are used to define a new crop area, applied to all seedlings on the plate (bottom). Click here to view larger image.
Discussion
Accurate phenotypic observation is crucial for understanding the manifestations of gene function within an organism. One way to acquire phenotypic information is through the capture of high-resolution image data. The scanner-based platform developed has enabled collection of many images (200 images/scan period) at high-resolution (4,800 dpi) over a number of hours. Additionally, this platform is easily adapted to a variety of lab and classroom environments due to the flexibility of the VueScan software to run thousands of different scanners using a common interface18.
The method presented here fills a void in high-throughput image capture that extends from large scale phenotyping facilities and automated systems implementable in a single laboratory. The high-throughput platforms currently available tend to use specialized imaging hardware, including cameras mounted on robotic supports, to capture high-resolution images of primarily above ground plant tissues (e.g. Centre for Plant Integrative Technology and the Scanalyzer HTS by LemnaTec)20,21. Specialized imaging systems using X-ray and MRI technologies have also been developed to image below ground tissues with remarkable resolution as they grow in the soil environment (e.g. Centre for Plant Integrative Technology) 11,22,23. This development of more specialized technology is generally at the cost of throughput, making dynamic phenotypic studies more difficult. Importantly, the cost and infrastructure needs for these high-end platforms make them mostly unfeasible for implementation in smaller laboratories.
Platforms have also been developed which use more standard image capture technology and are well suited to the measurement of dynamic responses such as the root response to a gravity stimulus. For example, CCD cameras have been used to capture individual seedling responses to light and gravity at high spatial and temporal resolution1,8,12. Other systems have been developed allowing measurement of root tip orientation of multiple roots from a single image (e.g. RootTipMulti by the iPlant Collaborative) 17,24. In the former case, throughput is relatively low given that only one seedling is imaged by each camera at a time, while in the latter case throughput is higher, but generally at the cost of resolution.
The procedure outlined in this paper presents a platform for capturing high-resolution images in high throughput with equipment and software that are readily available and relatively affordable. Using this setup, 1,080 individual root responses can be collected each week in a single lab equipped with a bank of six scanners. In 15 months of collecting an average of 864 individual responses per week, a total of 41,625 seedlings were scanned for a genomics study. About 15% of the individual collections failed due to setup error, network failure or equipment malfunction. Another 22% responses failed due to lack of germination or insufficient root growth to elicit a growth response. The final data set consists of 27,475 individual seedling responses to a gravity stimulus from 163 recombinant inbred lines plus 99 near isogenic lines. The data were collected in a single laboratory, making this a very high-throughput approach. Even given that the equipment used for acquisition is relatively inexpensive, it has functioned reliably for over two years even with heavy usage.
While this protocol has been very useful for the research aims of this group, some limitations still exist. Because of the throughput of about 50 GB of uncompressed image data per day, it was apparent that a large amount of space was needed to house images unless effective compression schemes could be developed. The storage problem was temporarily solved by purchasing external hard drives for each computer. In addition, two 10 TB network associated storage devices were purchased. Later, compression algorithms were developed, as described above, which can help reduce the data size by up to 60% (Figure 8). It is important to note that the speed at which data can be saved to a network associated storage device is dependent on the speed of the network connection. Compression schemes have also been constrained due to the desire to prevent loss of image data.
Other limitations specific to a scanner-based imaging system are also being considered. For example, in a scanner-based approach seedlings are exposed to high intensity light in the white and potentially infrared ranges during each scan. This likely affects seedling growth, though seedlings can still be observed to undergo robust responses to a gravity stimulus (Figure 7). A future improvement could involve programming scanners such that only infrared LEDs are active. An area in active development is creation of analysis algorithms well matched to the resolution and throughput of these image data. The large data set generated using this scanner-based method has been ideal for development of robust tools for high-throughput phenotyping of seedling images. The compression algorithm employed on these images shown in Figure 7 supports the claim that they are amenable to image analysis applications. Additionally, the images generated can be analyzed by the previously published algorithm, RootTrace17,24, if they are collected at lower resolution (less than 1,200 dpi), and individual seedlings are segmented from the image using the compression algorithm described above before analysis. Root growth data could be extracted from images reduced to 1,200 dpi while tip angle data could be extracted from images reduced to 900 dpi (unpublished observation).
The procedure outlined in this paper fits into its own niche in the world of root imaging in that it is high throughput and high resolution while still being relatively affordable. An additional benefit of this approach is that it can easily be customized to accommodate the imaging needs of a particular research group.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was funded by a grant from the National Science Foundation (award number IOS-1031416) and has been conducted in collaboration with Nathan Miller, Logan Johnson and Edgar Spalding of the University of Wisconsin and Brian Bockelman, Carl Lundstedt and David Swanson of the University of Nebraska's Holland Computing Center.
Materials
| Epson Perfection V700 Photo Scanners | Epson | B11B178011 | – |
| Plexiglas Scanner Template | – | – | Custom made. See Figure 2. |
| Smart Strap Bungee Cords | SmartStraps | Wal-Mart 1079478 | |
| Brinks Digital Outdoor Timers | Brinks | Wal-Mart 42-1014-2 | |
| VueScan Software | Hamrick Software | http://www.hamrick.com | |
| Segmentation Software | Chris Wentworth, Doane College | https://sites.google.com/a/doane.edu/compphy-doane/projects/root-gravitropism/image-segmentation | |
| 3M Micropore Tape | Fisher Scientific | 19-061-655 | – |
| Holding racks | – | – | Custom made by gluing two cookie racks together. |
Referências
- Miller, N. D., Brooks, T. L. D., Assadi, A. H., Spalding, E. P. Detection of a gravitropism phenotype in glutamate receptor-like 3.3 mutants of Arabidopsis thaliana using machine vision and computation. Genética. 186, 585-593 (2010).
- Clack, N. G. Automated Tracking of Whiskers in Videos of Head Fixed Rodents. PLoS Comp. Biol. 8, (2012).
- Lussier, Y. A., Liu, Y. Computational approaches to phenotyping: high-throughput phenomics. Proc. Am. Thoracic Soc. 4, 18-25 (2007).
- Houle, D. Colloquium Paper: Numbering the hairs on our heads: The shared challenge and promise of phenomics. Proc. Natl. Acad. Sci. U.S.A. 107, 1793-1799 (2009).
- Elwell, A. L., Gronwall, D. S., Miller, N. D., Spalding, E. P., L, T. D. B. Separating parental environment from seed size effects on next generation growth and development in Arabidopsis. Plant Cell Env. 34, 291-301 (2011).
- Silk, W. K. Quantitative Descriptions of Development. Ann. Rev. Plant Physiol. 35, 479-518 (1984).
- Cronin, C. J., Feng, Z., Schafer, W. R. Automated imaging of C. elegans behavior. Methods Mol. Biol. 351, 241-251 (2006).
- Miller, N. D., Parks, B. M., Spalding, E. P. Computer-vision analysis of seedling responses to light and gravity. Plant J. 52, 374-381 (2007).
- Iyer-Pascuzzi, A. S. Imaging and Analysis Platform for Automatic Phenotyping and Trait Ranking of Plant Root Systems. Plant Physiol. 152, 1148-1157 (2010).
- Houle, D., Mezey, J., Galpern, P., Carter, A. Automated measurement of Drosophila wings. BMC Evol. Biol. 3, 25 (2003).
- Jahnke, S. Combined MRI-PET dissects dynamic changes in plant structures and functions. Plant J. 59, 634-644 (2009).
- Durham Brooks, T. L., Miller, N. D., Spalding, E. P. Plasticity of Arabidopsis Root Gravitropism throughout a Multidimensional Condition Space Quantified by Automated Image Analysis. Plant Physiol. 152, 206-216 (2010).
- Perrin, R. M. Gravity signal transduction in primary roots. Ann. Botany. 96, 737-743 (2005).
- Strohm, A. K., Baldwin, K. L., Masson, P. H. Molecular mechanisms of root gravity sensing and signal transduction. Dev. Biol. 1, 276-285 (2012).
- Harrison, B. R., Masson, P. H. ARL2, ARG1 and PIN3 define a gravity signal transduction pathway in root statocytes. Plant J. 53, 380-392 (2007).
- Swarup, R., Bennett, M. J., Beeckman, T. . Root Development. , 157-174 .
- French, A., Ubeda-Tomás, S., Holman, T. J., Bennett, M. J., Pridmore, T. High-throughput quantification of root growth using a novel image-analysis tool. Plant Physiol. 150, 1784-1795 (2009).
- Granier, C. PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. , 169-623 (2006).
- Walter, A. Dynamics of seedling growth acclimation towards altered light conditions can be quantified via GROWSCREEN: a setup and procedure designed for rapid optical phenotyping of different plant species. New Phytol. 174, 447-455 (2007).
- Gregory, P. J. Non-invasive imaging of roots with high resolution X-ray micro-tomography. Plant Soil. , 255-351 (2003).
- Pierret, A., Kirby, M., Moran, C. Simultaneous X-ray imaging of plant root growth and water uptake in thin-slab systems. Plant Soil. 255, 361-373 (2003).
- Naeem, A., French, A. P., Wells, D. M., Pridmore, T. P. High-throughput feature counting and measurement of roots. Bioinformatics. 27, 1337-1338 (2011).

