Two-photon Calcium Imaging in Mice Navigating a Virtual Reality Environment
Summary
Here we describe the experimental procedures involved in two-photon imaging of mouse cortex during behavior in a virtual reality environment.
Abstract
In recent years, two-photon imaging has become an invaluable tool in neuroscience, as it allows for chronic measurement of the activity of genetically identified cells during behavior1-6. Here we describe methods to perform two-photon imaging in mouse cortex while the animal navigates a virtual reality environment. We focus on the aspects of the experimental procedures that are key to imaging in a behaving animal in a brightly lit virtual environment. The key problems that arise in this experimental setup that we here address are: minimizing brain motion related artifacts, minimizing light leak from the virtual reality projection system, and minimizing laser induced tissue damage. We also provide sample software to control the virtual reality environment and to do pupil tracking. With these procedures and resources it should be possible to convert a conventional two-photon microscope for use in behaving mice.
Introduction
Two-photon imaging of calcium indicators (genetically encoded like GCaMP57 or R-GECO8, or synthetic dyes like OGB or Fluo4) has emerged as a powerful method of measuring neuronal activity in behaving mice1-6. It enables the simultaneous measurement of the activity of hundreds of cells at near-single action potential resolution, up to approximately 800 µm below the brain surface9,10. Moreover, using genetically encoded calcium indicators (GECIs) neuronal activity can be measured chronically5,11,12, and in genetically defined cell types13. Together, these methods provide a degree of temporal and spatial resolution that open up a multitude of new possibilities in the study of neuronal computation in vivo.
Surgical intervention is necessary to expose and label the mouse brain for imaging. Cells are typically transfected using a recombinant adeno-associated virus (AAV) system for GECI delivery and a cranial window is implanted over the injection site to gain optical access to the brain. A head bar is then attached to the skull for head fixation under the two-photon microscope. The design and implementation of these steps is critical as most of the problems with awake imaging arise from instabilities in the preparation. Ideally the procedure described here should allow for chronic imaging of up to several months following the surgery.
To enable visually guided behavior during two-photon imaging, the head fixed mouse sits on an air supported spherical treadmill, which it can use to navigate a virtual reality environment. Locomotion of the mouse on the treadmill is coupled to movement in the virtual environment that is displayed on a toroidal screen surrounding the mouse14,15. Behavioral variables such as locomotion, visual stimulus, and pupil position can be recorded6.
We describe the procedures involved in chronic two-photon imaging in mice exploring a virtual reality environment. The key points addressed are: reduction of movement artifacts, reduction of light leak, maximization of the number of simultaneously recorded cells, and minimization of photo damage. We also provide details on setting up the air-supported treadmill, pupil tracking, and the virtual reality environment. The procedures described here can be used for imaging fluorescently labeled cell populations in head fixed mice in a potentially wide variety of behavioral paradigms.
Protocol
All animal procedures were approved and carried out in accordance with guidelines of the Veterinary Department of the Canton Basel-Stadt.
1. Hardware and Software Setup
- Two-photon scanning microscope setup:
- Use a pulsed infrared laser as an illumination source (pulse width <120 fsec).
- Use a scan head composed of an 8 or 12 kHz resonant scanner and a standard galvanometer. Note: This enables frame rates of 40 or 60 Hz at 750 x 400 pixels. A high frame rate is critical for minimizing brain motion induced image distortion. Moreover, fast resonant scanning results in a higher signal yield and reduces photobleaching16 and phototoxicity.
- Use a Piezo-electric high speed z-stepper to sequentially acquire time interleaved images at multiple depths. Note: This is optional and used to increase data yield at the expense of frame rate.
- Use a mechanical blanker to reduce laser exposure to the tissue. Adjust the width of the blanker depending on the amplitude of the resonant scanner. Note: Due to the sinusoidal nature of the scan path of a resonant scanner, the laser beam moves comparably slow at the scan turnaround points. To prevent damage of the tissue at these turnaround points where laser exposure is greatest, a mechanical blanker is introduced into the focal plane between the scan and tube lenses that stops the laser beam.
- For data acquisition use a high speed digitizer (800 MHz) in combination with a field-programmable gate array (FPGA). Note: The FPGA high-pass filters and bins the PMT signal into pixels. The combination of the high-pass filter on the FPGA and the effective low-pass filter on the PMT amplifier result in a band-pass filter centered roughly around the repetition rate of the laser (80 MHz). Sample FPGA code is provided as a supplement.
- Air-supported treadmill setup based on the design of Dombeck and colleagues2
- Connect the ball holder to an air supply line using an air supply tube of at least 10 mm inner diameter. Maximize the cross section of the ball holder air supply inlet and place ample amount of tubing between the main air supply and the ball holder. Note: Increased cross section and increased length of the air supply tubing reduces noise generation.
- Place a polystyrene foam ball (20 cm diameter) in a custom-designed and 3D-printed ball holder.
- If the behavioral paradigm only requires forward (and backward) locomotion, restrict movement of the ball to the horizontal (pitch) axis by inserting a pin (e.g. a 19 G hypodermic needle) on the side of the ball. Note: Typically animals adapt to the setup much quicker in this reduced rotation paradigm.
- Track movement of the ball using one or two optical computer mice to detect motion around two or all three axes, respectively. Note: The computer mice should ideally operate with an infrared diode for least interference with the imaging.
- Virtual reality environment setup
- Prepare the virtual reality environment. Note: A sample environment based on Panda3D, a freely available game engine, is provided as a supplement. The objects of the virtual environment are generated using Wings3D, an open source 3D modeler. The sample program reads position information on a data acquisition card and translates this into movement in the virtual environment. The code also performs the nonlinear transformation necessary for the environment to be projected onto a toroidal screen. The detailed construction of the toroidal screen has been described elsewhere14,17.
- Use LED projectors or LED backlit computer screens for displaying the virtual environment. Integrate a gating circuit for fast flickering of the LEDs during imaging. Note: An inherent problem in two-photon imaging during visual stimulation is light leak originating from the visual stimulation system, which can be reduced by shielding the objective. However, light leak from the visual stimulation system can be fully abolished by synchronizing the light output of the projector to the turnaround points of the resonant scanner (when data are not acquired), as is done with LED arenas used for visual stimulation during imaging in Drosophila18. This results in an effective high speed alternation of visual stimulation and data acquisition (see Figure 2). The flickering frequency (24 kHz) is orders of magnitude above the flicker fusion threshold, above which flickering is no longer perceived by the animal. The electronic circuit diagram of this modification to a commercial LED projector is shown in Figure 3.
- Pupil tracking
- Use a CMOS based video camera (30 fps) for pupil position tracking. Make sure that the camera does not contain an infrared blocking filter. Note: The pupil is visible at high contrast during recordings due to stray light from the two-photon excitation laser exiting through the eye. Pupil position and diameter is extracted in real-time using custom-programmed software based on the design of Sakatani and Isa19.
2. Injection of Genetic Calcium Indicator and Window Implant
The injection of a calcium indicator20 and the implantation of the cranial window21 are performed as previously described with the following additions:
- Inject the virus into the region of interest using a glass injection pipette backfilled with virus solution and connected to a pressure injection system. Adjust the pressure depending on the pipette inner-diameter, typically approx. 70-140 mbar, to inject approximately 100 nl of virus solution over the course of 1-2 min. Low volume injection pulses delivered at low frequency (0.05 sec pulses at 2 Hz) should be used in order to prevent overflow of the virus along the pipette during injection. Note: In order to monitor the injection volume, the pipette can be marked at small, evenly spaced lengths (e.g. 0.5 mm) under a stereo microscope. Displacement of the meniscus between the markings can then be used to estimate the injected volume.
- Several weeks post injection this should result in a bolus of densely labeled cells with a diameter of approximately 500 µm. Efficacy of GECI expression in the region of interest depends on the choice of GECI and promoter combination, as well as the infectious titer and serotype of the recombinant AAV used. Note: As a representative example, injection of AAV2/1-EF1α-GCaMP5 with a viral titer of 8 x 1012 GC/ml into primary visual cortex of a mouse results in satisfactory expression at approximately two weeks post injection (Figure 1).
- The head bar should not obstruct the visual field of the mouse, but should be rigid enough to allow for stable imaging during behavior. This typically requires two points of attachment on either side of the head bar (see Figure 1). To firmly attach the head bar to the skull, make sure to maximize and dry the area of exposed skull where the head bar will be attached. Additionally, using a scalpel or a syringe, scratch the skull to create superficial grooves thereby increasing the surface area for gluing. Cover the bone with cyanoacrylate tissue adhesive before attaching the head bar with dental cement.
3. Experimental Procedure
- Two to three weeks following virus injection, check the cranial window implant for clarity and expression under epifluorescence illumination. Clearly visible and sharply defined boundaries of superficial blood vessels are a good indication that the window implant is suitable for imaging (see Figure 1).
- Measure and adjust laser power to a value below 50 mW at the sample.
- Turn on the airflow to the spherical treadmill.
- For head fixation under the microscope, briefly anesthetize the animal with isoflurane to reduce stress. Place the animal into an isoflurane anesthesia container and wait until the animal stops moving (approximately 10 sec). Remove the animal and immediately head fix it under the microscope.
- Position the visual display system as close as possible to the animal to still allow for access to the animal. The less visual contact the animal has with the experimenter, the less stressed the animal will be during the initial phase of the experiment.
- Make sure the cranial window implant is free of dust and dirt. Clean with water if necessary.
- Apply clear ultrasound gel as immersion medium between the cranial window and water-immersion objective. This solves problems of slow image degradation due to water evaporation or leakage and alleviates the need to mount a cone on the head bar to hold water. Note: Centrifuge the gel before usage to remove all air bubbles, and take care not to create any air bubbles during application of the gel. Air bubbles can severely degrade image quality.
- Wrap a piece of black tape around the objective and head bar for light shielding.
- Move the virtual environment arena to its final position and adjust the video camera for pupil tracking.
- Set the laser blanker to match the scan amplitude used during the experiment.
- Start recording data.
Representative Results
The image quality in two-photon calcium imaging of cell populations labeled with a GECI largely depends on the quality of the cranial window implant. Two weeks following virus injection the cranial window should be inspected for clarity. There should be no granulation tissue or bone regrowth visible (Figure 1A). Moreover, the pattern of superficial blood vessels should remain unchanged and boundaries of the vasculature should be sharply defined. At the same time, GECI expression can also be checked. Boli of labeled cells should be clearly visible under an epifluorescence microscope (Figure 1B).
Ideally the preparation is stable enough such that distortions of the two-photon images induced by locomotion of the mouse are not visible in the image after frame registration (Figure 2). The scan path of the resonant scanner is sinusoidal resulting in nonlinear image distortion. During the turnaround points data are not acquired due to image stretching in this part of the scan path. If the light source of the visual stimulation system, here the LEDs of the projector, is correctly timed to the turnaround points, light leak should only be apparent during this time (Figure 2). The flickering frequency of the light source has to be twice the scan frequency. Key to making this work are the fast rise and fall times of the LEDs and controlling electronics (Figure 3).
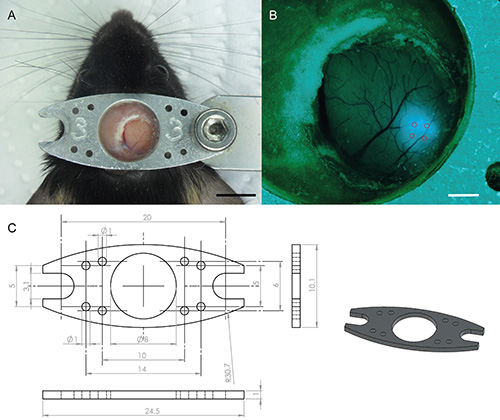
Figure 1. Top view of a cranial window in a head fixed mouse two weeks after surgery. (A) The head bar is optimized in weight and shape to increase stability for behavior during imaging experiments, while reducing burden on the animal. The head bar is made from aluminum and has a thickness of 1 mm. Scale bar is 5 mm. (B) Epifluorescence image of GCaMP5 expression after four injections (AAV2/1-EF1α-GCaMP5) of approximately 80 nl. The injection depth is approximately 400 µm. Scale bar is 1 mm. (C) Schematics of the head bar. All measurements in mm. Click here to view larger image.
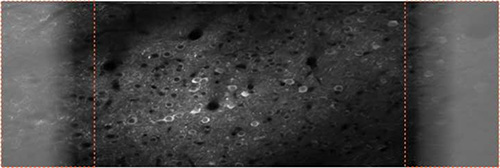
Figure 2. Improving signal to noise by reducing light leak. The LED light source in the projector of the virtual reality environment is synchronized to the resonant scanner to turn on only during the scan turnaround periods when data are not recorded (orange boxes). The laser blanker was removed from the microscope for this example to illustrate the effect of light leak resulting from visual stimulation. The image (average of 8 sec of data) was taken in mouse visual cortex transfected with AAV2/1-EF1α-GCaMP5. Click here to view larger image.
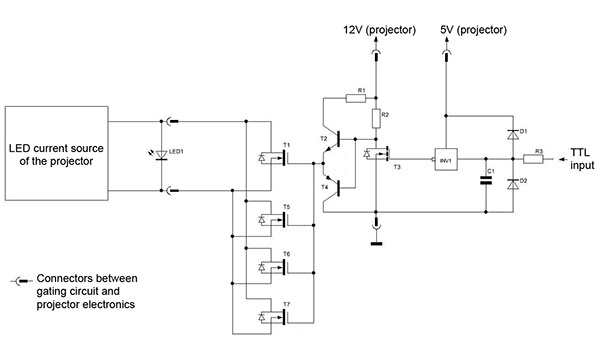
Figure 3. Circuit diagram of the electronics used to flicker a conventional LED projector. The gating circuit is driven by a TTL input to synchronize fast flickering with the resonant scanner. C1: capacitor 1nF; D1, D2: diodes 1N4148; INV1: Inverter HEF40106 (1 of 6 used); LED1 – LED of the beamer; R1: resistor 47 Ω; R2: resistor 2.2 kΩ; R3: resistor 100 Ω; T1, T5, T6, T7: transistors IF9321; T2: transistor BC846; T3: transistor BC170; T4: transistor BC856. Note that this circuit has to be repeated for all three color channels (red, green, blue) of the projector. Click here to view larger image.
Discussion
The key to the success of behavioral two-photon imaging is the stability of the preparation in two ways:
- Over the course of days post window implantation, inflammatory responses of the tissue can lead to enhancement of the formation of granulation tissue and cartilage that will hinder or even prevent imaging.
- During the experiment the brain has to be stable enough to prevent motion artifacts from corrupting neural activity related fluorescence signal.
To keep the inflammatory immune response to a minimum the cranial window implantation surgery should be carried out as quickly as possible and direct damage to the brain should be avoided at all costs. Of paramount importance is to ensure that the cranial window implant remains airtight for the entire duration of the experiment to prevent inflammation.
To minimize brain motion related imaging artifacts take care not to damage or remove the dura mater during the surgery. Remaining locomotion induced brain motion can be corrected for when motion is confined to the plane parallel to the imaging plane (the x/y plane). The maximum tolerable standard deviation of motion in x/y is roughly 5 µm. For motion along the z-axis this value is roughly 1 µm, such that the extent of the movement is smaller than the resolution limit of the microscope.
The low speed of the resonant scanner at the scan path turnaround points can lead to laser induced tissue damage. To prevent this, the laser can be blocked at the turnarounds using a blanker in the scan telescope. If this blanker is set correctly and average laser power under the objective is kept below 50 mW, imaging is possible for extended periods of time over multiple days without causing any visible laser damage. An alternative method to a mechanical blanking of the laser is to use a fast Pockels cell to reduce laser power at the turnarounds. This method however has the disadvantage that Pockels cells typically introduce undesirable beam distortions.
The aptitude of head fixed rodents to navigate virtual environments has made it possible to chronically image populations of cells in cortex of mice performing a wide range of behavioral tasks3,4,6,22. However, it is worthwhile to note that head-fixation inherently constrains behavior and locomotion on a spherical treadmill is in many ways not equivalent to unrestrained locomotion or normal behavior.
In the future, the strength of the virtual reality imaging approach described here will likely be the combination of calcium imaging with targeted optogenetic stimulation during behavior. With this it would be possible to optically manipulate presynaptic cells and image their post-synaptic targets while monitoring the resultant behavioral changes. In addition, as calcium indicators freely diffuse within the cell, it will also be possible to measure activity selectively in subcellular compartments during behavior. Due to movement induced brain motion, such recordings have been limited by the difficulty to obtain stable recordings.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Friedrich Miescher Institute for Biomedical Research, the Max Planck Society, and the Human Frontiers Science Program.
Materials
| cover slips (d = 3-5 mm) | Menzel | window implant | |
| InSight DeepSee laser | Spectra-Physics | microscope | |
| 12kHz resonance scanner | Cambridge Technology | G1-003-30026 | microscope |
| Galvometer | Cambridge Technology | G6215H | microscope |
| Digitizer | National Instruments | NI 5772 | microscope |
| FPGA | National Instruments | PXIe 7965R | microscope |
| Acquisition card | National Instruments | PCIe 6363 | microscope |
| Emission filter 525/50 | Semrock | FF03-525/50-25 | microscope |
| Piezo-electric z-drive | Physikinstrumente | P-726.1CD | microscope |
| Controller for piezo-electric drive | Physikinstrumente | E665 LVPZT | microscope |
| Objective 16x, 0.8NA | Nikon | CFI75 | microscope |
| Current amplifier | Femto | DHPCA-100 | microscope |
| Photomultiplier tube | Hamamatsu | microscope | |
| USB Camera without IR filter | ImagingSource | DMK22BUC03 | pupil tracking |
| Objective 50 mm | ImagingSource | M5018-MP | pupil tracking |
| Macro adapter rings | ImagingSource | LAexSet | pupil tracking |
| Optical computer mouse | Logitech | G500 | motion tracking |
| Styrofoam ball 20 cm | e.g. idee-shop.de | 08797.00.15 | virtual environment |
| LED projector | Samsung | SP-F10M | virtual environment |
| Acquisition card | National Instruments | NI 6009 | virtual environment |
| Panda3D game engine | www.panda3d.org | virtual environment | |
| Numpy library for Python | www.scipy.org | virtual environment | |
| Scipy library for Python | www.scipy.org | virtual environment | |
| NI-DAQmx driver | National Instruments | www.ni.com | virtual environment |
| Ultrasound gel | Dahlhausen | 5701.0342.10 | imaging |
Referências
- Helmchen, F., Fee, M. S., Tank, D. W., Denk, W. A Miniature Head-Mounted Two-Photon MicroscopeHigh-Resolution Brain Imaging in Freely Moving Animals. Neuron. 31 (6), 903-912 (2001).
- Dombeck, D. A., Khabbaz, A. N., Collman, F., Adelman, T. L., Tank, D. W. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron. 56, 43-57 (2007).
- Dombeck, D. A., Harvey, C. D., Tian, L., Looger, L. L., Tank, D. W. Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat. Neurosci. 13 (11), 1433-1440 (2010).
- Harvey, C. D., Coen, P., Tank, D. W. Choice-specific sequences in parietal cortex during a virtual-navigation decision task. Nature. 484 (7395), 62-68 (2012).
- Huber, D., Gutnisky, D. A. Multiple dynamic representations in the motor cortex during sensorimotor learning. Nature. 484 (7395), 473-478 (2012).
- Keller, G. B., Bonhoeffer, T., Hübener, M. Sensorimotor mismatch signals in primary visual cortex of the behaving mouse. Neuron. 74 (5), 809-815 (2012).
- Akerboom, J., Chen, T. -. W. Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. J. Neurosci. 32 (40), 13819-13840 (2012).
- Zhao, Y., Araki, S. An expanded palette of genetically encoded Ca2+ indicators. Science. 333 (6051), 1888-1891 (2011).
- Mittmann, W., Wallace, D. J. Two-photon calcium imaging of evoked activity from L5 somatosensory neurons in vivo. Nat. Neurosci. 14 (8), 1089-1893 (2011).
- Katona, G., Szalay, G. Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat. Methods. 9 (2), 201-208 (2012).
- Mank, M., Santos, A. F. A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat. Methods. 5 (9), 805-811 (2008).
- Margolis, D. J., Lütcke, H. Reorganization of cortical population activity imaged throughout long-term sensory deprivation. Nat. Neurosci. 15 (11), 1539-1546 (2012).
- Zariwala, H. A., Borghuis, B. G. A Cre-dependent GCaMP3 reporter mouse for neuronal imaging in vivo. J. Neurosci. 32 (9), 3131-3141 (2012).
- Harvey, C. D., Collman, F., Dombeck, D. A., Tank, D. W. Intracellular dynamics of hippocampal place cells during virtual navigation. Nature. 461 (7266), 941-946 (2009).
- Hölscher, C., Schnee, A., Dahmen, H., Setia, L., Mallot, H. A. Rats are able to navigate in virtual environments. J. Exp. Biol. 208, 561-5519 (2005).
- Borlinghaus, R. T. MRT letter: high speed scanning has the potential to increase fluorescence yield and to reduce photobleaching). Microsc. Res. Tech. 69 (9), 689-692 (2006).
- Reiff, D. F., Plett, J., Mank, M., Griesbeck, O., Borst, A. Virtual Reality for Mice, mousevr.blogspot.com. Nat. Neurosci. 13, 973-978 (2010).
- Sakatani, T., Isa, T. Quantitative analysis of spontaneous saccade-like rapid eye movements in C57BL/6 mice. Neurosci. Res. 58, 324-331 (2007).
- Golshani, P., Portera-Cailliau, C. In vivo 2-photon calcium imaging in layer 2/3 of mice. J. Vis. Exp. (13), (2008).
- Holtmaat, A., Bonhoeffer, T. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat. Protoc. 4 (8), 1128-1144 (2009).
- Schmidt-Hieber, C., Häusser, M. Cellular mechanisms of spatial navigation in the medial entorhinal cortex. Nat. Neurosci. 16 (3), 325-331 (2013).

