Laser-induced Breakdown Spectroscopy: A New Approach for Nanoparticle’s Mapping and Quantification in Organ Tissue
Summary
Laser-induced breakdown spectroscopy performed on thin organ and tumor tissue successfully detected natural elements and artificially injected gadolinium (Gd), issued from Gd-based nanoparticles. Images of chemical elements reached a resolution of 100 μm and quantitative sub-mM sensitivity. The compatibility of the setup with standard optical microscopy emphasizes its potential to provide multiple images of a same biological tissue.
Abstract
Emission spectroscopy of laser-induced plasma was applied to elemental analysis of biological samples. Laser-induced breakdown spectroscopy (LIBS) performed on thin sections of rodent tissues: kidneys and tumor, allows the detection of inorganic elements such as (i) Na, Ca, Cu, Mg, P, and Fe, naturally present in the body and (ii) Si and Gd, detected after the injection of gadolinium-based nanoparticles. The animals were euthanized 1 to 24 hr after intravenous injection of particles. A two-dimensional scan of the sample, performed using a motorized micrometric 3D-stage, allowed the infrared laser beam exploring the surface with a lateral resolution less than 100 μm. Quantitative chemical images of Gd element inside the organ were obtained with sub-mM sensitivity. LIBS offers a simple and robust method to study the distribution of inorganic materials without any specific labeling. Moreover, the compatibility of the setup with standard optical microscopy emphasizes its potential to provide multiple images of the same biological tissue with different types of response: elemental, molecular, or cellular.
Introduction
The wide development of nanoparticles for biological applications urged the parallel improvement of analytical techniques for their quantification and imaging in biological samples. Usually the detection and the mapping of the nanoparticles in organs are made by fluorescence or confocal microscopy. Unfortunately these methods require the labeling of the nanoparticles by a near infrared dye that can modify the biodistribution of the nanoparticles, especially for very small nanoparticles due to its hydrophobic properties. The detection of labeled nanoparticles, and especially the very small nanoparticles (size < 10 nm), might thus interfere with their biodistribution at the whole body scale but also at the tissue and cell levels. The development of new devices able to detect nanoparticles without any labeling offers new possibilities for the study of their behavior and kinetics. Moreover, the role of trace elements such as iron and copper in brain illnesses and neurodegenerative diseases such as Alzheimer1, Menkes2,3, or Wilson4 suggest the interest to study and localize these elements in tissues.
Various techniques have been used to provide elemental mapping or microanalysis of different materials. In their review paper published in 2006, R. Lobinski et al. provided an overview of available standard techniques for elemental microanalysis in biological environment, one of the most challenging environments for analytical sciences5. The electron microprobe, which consists of energy dispersive X-ray microanalysis in a transmission electron microscope, can be applied to numerous studies if the element concentration is sufficient (>100–1,000 μg/g). To reach lower detection limits, the following techniques have been used:
- ion beam microprobe using particle induced X-ray emission μ-PIXE (1–10 μg/g)6
- synchrotron radiation microanalysis μ-SXRF (0.1–1 μg/g)7
- secondary ion mass spectrometry SIMS (0.1 μg/g)8
- laser ablation inductively coupled mass spectrometry LA-ICP-MS (down to 0.01 μg/g)9,10
The above-mentioned techniques provide micrometric resolution as shown in the Table 1 extracted from Lobinski et al.
3D reconstruction of serial 2D investigations could also be proposed for the reconstruction of deeper tissues11. However, all devices and systems require both qualified professionals, moderate to highly expensive equipment and long-lasting experiments (typically more than 4 hr for a 100 µm x 100 µm for µ-SXRF and 10 mm x 10 mm for LA-ICP-MS)12. Altogether, these requirements make elemental microanalysis very restricting and incompatible with conventional optical imaging systems, fluorescence microscopy or nonlinear microscopy. Another point that we can mention here is that the quantitative measurement capability is still quite limited and depends on the availability of matrix-matched laboratory standards. The further generalization of the use of elemental microanalysis in industry processes, geology, biology and other domains of applications will generate significant conceptual and technological breakthroughs.
The purpose of the present manuscript is to propose solutions for quantitative elemental mapping (or elemental microanalysis) in biological tissues with a tabletop instrumentation fully compatible with conventional optical microscopy. Our approach is based on the laser-induced breakdown spectroscopy (LIBS technology). In LIBS, a laser pulse is focused on the sample of interest to create the breakdown and spark of the material. The atomic radiation emitted in the plasma is subsequently analyzed by a spectrometer and the elemental concentrations can be retrieved with calibration measurements performed beforehand13,14. The advantages of LIBS include sensitivity (µg/g for almost all the elements), compactness, very basic sample preparation, absence of contact with the sample, instantaneous response and precisely localized (micro) surface analysis. However, the application of tissue chemical imaging remains challenging since the laser ablation of tissue must be finely controlled to perform maps with high spatial resolution together with sensitivity in the µg/g range15,16.
With such solution, the adjunction of tracers or labeling agents is not needed, which allows detecting inorganic elements directly in their native environment in biological tissues. The LIBS instrument developed in our laboratory offers a current resolution inferior to 100 µm with an estimated sensitivity for Gd below 35 µg/g, equivalent to 0.1 mM 16, which allows the mapping of large samples (>1 cm2) within 30 min. In addition, homemade software facilitates the acquisition and exploitation of the data. This instrument is used to detect, map, and quantify the tissue distribution of gadolinium (Gd)-based nanoparticles17–18 in kidneys and tumor samples from small animals, 1 to 24 hr after intravenous injection of the particles (size <5 nm). Inorganic elements, which are intrinsically contained in a biological tissue, such as Fe, Ca, Na, and P, have also been detected and imaged.
Protocol
1. Biological Sample Preparation
All the experiments described in this study were approved by the Animal Care and Use Committee of the CECCAPP (Lyon, France) (authorization #LYONSUD_2012_004), and the experiments were carried out under the supervision of authorized individuals (L. Sancey, DDPP authorization #38 05 32).
- Add 1 ml of H2O to 100 µmol of Gadolinium (Gd)-based nanoparticles, wait 15 min, and add 20 µl of HEPES 50 mM, NaCl 1.325 M, CaCl2 20 mM to 100 µl of H2O and 80 µl of the primary solution of Gd-based nanoparticles to obtain a 200 µl solution at 40 mM ready-to-inject (City: Villeurbanne, at lab).
- Inject the 200 µl Gd-based nanoparticles solution intravenously into anesthetized tumor-bearing rodents (City: Lyon Sud (Oullins), 15 km from the lab).
- 1-24 hr after injection, sacrifice the mice and put the biological samples into isopentane cooled by liquid nitrogen. Store the samples at – 80 °C (City: Lyon Sud (Oullins), 15 km from the lab).
- Slice the sample (City: usually Grenoble, 100 km from the lab; I will try to have an access in Lyon Sud) in 100 µm-thick slides and put the biological slides on specific plastic dishes (Petri dish). Store at -80 °C.
Note: Plastic dishes are basically a very pure polymer sample. They are used to avoid interference with elements contained in the tissue.
2. Sample Preparation for Calibration
- Prepare vials with increasing doses of Gd-based nanoparticle into water (0 nM, 100 nM, 500 nM, 1 µM, 5 µM, 10 µM, 50 µM, 100 µM, 500 µM, 1 mM, and 5 mM).
- Put a 5 µl-drop of each solution regularly spaced by 3 mm on the Petri dish.
- Dry at room temperature for 20 min.
3. LIBS Experiment
- Initializing the LIBS Setup
- Laser Settings. After switching on the instruments, wait 10-20 min for laser pulse energy stabilization and cooling down of the ICCD camera to -20 °C. Adjust the pulse energy with the attenuator.
Note: The optimum laser parameters for mapping tissues are 5 nsec pulse duration, 1,064 nm wavelength, and pulse energy of about 4 mJ. The laser uses is a typical Nd:YAG nanosecond laser. - LIBS Settings. Set the laser focusing position (with respect to the sample surface) to obtain the smaller crater diameter (about 50 µm or less).
Note: This corresponds to a laser pulse focalization 100 µm below the sample surface. - Spectrometer Settings. Use the Czerny-Turner spectrometer combined with a 1,200 lines/mm grating and a high temporal resolution ICCD camera. Control all these devices by computer. Set the input slit value to 40 µm. Set the spectral range regarding the element to be analyzed. Use the spectral range covering 325 to 355 nm to detect Gd with high sensitivity, as well as Na, Cu and Ca. Set the ICCD parameters with a delay of 300 nsec, a gate of 2 µsec, and a gain of 200.
- Laser Settings. After switching on the instruments, wait 10-20 min for laser pulse energy stabilization and cooling down of the ICCD camera to -20 °C. Adjust the pulse energy with the attenuator.
- Mapping Measurement
- Place the biological sample on the LIBS motorized sample holder.
- Adjust the height of the sample according to the laser focus position.
- Take a high resolution photograph of the sample slice.
- Set the mapping module of the LIBS acquisition software to perform a map with typically 100 x 100 measurement points spaced by a resolution of about 100 µm.
- Start the acquisition. From this point automate everything; the moving sequence as well as the spectrum recording and saving. 40 min are required for a mapping of 10,000 points (equivalent to 1 cm2 for 100 µm resolution). Regroup all the recorded spectra into a same file.
- When finished, take a second photograph of the sample slice
- Calibration Measurement
With the same experimental parameters, measure the calibration samples (for preparation details, see section 2). Perform a map or record 25 spectra (obtained from measurement sites in the center part of the drop) in each of the calibration drops.
4. LIBS Spectrum Analysis: Constructing of Chemical Images
- Record all LIBS spectra from the tissue mapping and load them in the LIBS software analysis. Subtract the baseline for each spectrum and construct the chemical images with relative intensity scale using a false color.
Note: An algorithm retrieves specific line intensities, such as Gd, Cu, Na, or Ca. - Perform the same operation on the spectra measured from the calibrated sample to allow the calibration curves calculation (relation between intensity and concentration) and build a quantitative map or image for Gd (or other element of interest).
- Apply the adequate treatments to the chemical images, such as interpolation or smoothing. Save the intensity or concentration maps in image format (bitmap).
Representative Results
As shown in Figure 1, the beam of a Nd:YAG laser in the fundamental wavelength of 1,064 nm was focused vertically down on the tissue slice by a quartz lens of 50 mm focal distance. The pulse energy was 4 mJ and the repetition rate 10 Hz. In order to avoid the generation of plasma in air, the laser beam was focused around 100 µm under the surface of the sample. No air plasma was observed in this condition. During the experiments, the sample was moved by a step motor in order to generate one plasma in only one position on the sample (single shot). A microscopic observation of the craters generated on the substrate showed a crater diameter lower than 50 µm. The generated plasma was imaged by a couple of lens onto the entrance of an optical fiber connected at the other end to a Czerny-Tuner spectrograph equipped with a grating of 1,200 lines per mm (blazed at 300 nm). An ICCD camera was mounted on the output focal plane of the spectrograph to record the spectrum. During the measurements, the height of the sample surface (with regard to the laser focus position) is monitored using trigonometric method with an obliquely incident beam from a low power cw laser diode on the sample surface and a CCD monitor camera. The ICCD was triggered by the laser Q-switch and the 400 nsec and 2 µsec were set respectively for the delay and gate parameters. During a mapping, the sample was shifted by an offset of 100 µm after each laser shot. One spectrum was recorded for each laser shot. Homemade software developed in the LabVIEW environment controlled the entire equipment and allowed to perform automated sequence to scan the area of interest of the tissue sample with specific lateral resolution.
Example of single shot spectra, recorded on different regions of a kidney tissue after IV injection of Gd-based nanoparticles is shown in Figure 2. The nanoparticles were synthesized as described in Mignot et al17. Briefly, the nanoparticles are composed of a polysiloxane matrix, holding DOTAGA-Gd3+ chelates on their surface. They are developed for (i) multi-modal imaging as they can be used in MRI, nuclear imaging and fluorescence imaging, separately or simultaneously, and (ii) therapeutic function as radiosensitizer (i.e. increasing locally the efficiency of the radiotherapy)18. The spectral range used (286 to 320 nm) allows detecting different elements such as Gd, Ca, Fe, Si, and Al. The blue spectrum was recorded in the central region of the kidney (the medulla), the red spectrum corresponded to the kidney membrane (capsule) and the green spectrum to the peripheral area (cortex). It is also noteworthy that the Gd, Si, Al, Ca, and Fe intensities are widely variable at different regions, suggesting a large heterogeneity of these elements concentrations within the tissue. Basically Gd and Si were containing in the Gd-based nanoparticles, Fe was specific for the blood vessels, Ca from the biological sample itself.
The tissue slice samples were analyzed spot by spot, moving the sample in the X and Y position. Lateral resolution was close to 100 µm. One single spectrum was recorded for each position (single shot measurement). Line intensities were extracted from the spectra using a background subtraction. The chemical images were then constructed from these intensities. An example is shown in Figure 3 for Gd, Si and Fe. Lines used to build these chemical images are shown in Table 2. The analysis was correlated to the colored photography of the biological sample before the experiment to improve the localization and mapping of the different elements. In the kidney, the Gd and Si signals were co-localized, but this co-localization did not fit with the distribution of the Fe, indicating a specific distribution.
Similar results were obtained with tumor samples. An example of measurement on SQ20B tumor tissue is shown in Figure 4. In this case, the chemical map of Gd, shown with a false color scale, has been superposed to the natural light picture. For this measurement, quantitative analysis has been performed to retrieve the local gadolinium concentration (see the procedure in the protocol section). For this experiment, the nanoparticles were administered directly into the tumor, which was removed 1 hr after injection and sliced for the analysis. As seen in Figure 4, the particles diffused in the tissue from the injection point in the tumor center to the periphery. 1 hr after the injection, approximately half of the tumor volume contained some particles. Such a mapping can help to provide information about the therapeutic protocol. For an optimal efficacy of the treatment, the nanoparticles should diffuse within the entire tumor; after 1 hr, only half of the tumor contained some particles, suggesting that a longer diffusion time would be required for an optimal diffusion and thus effective treatment.
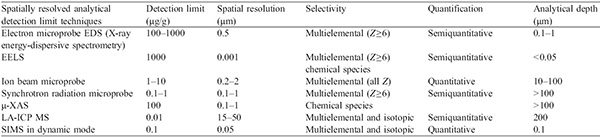
Table 1. Main spatially resolved analytical techniques for chemical elemental imaging. (Lobinski et al.5). Please click here to view a larger version of this table.
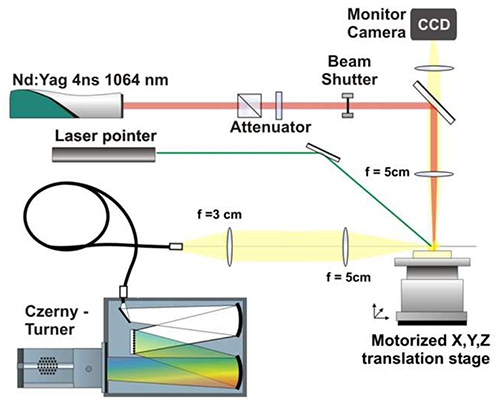
Figure 1. Schematic presentation of the used LIBS experimental setup. (Motto-Ros et al.15).
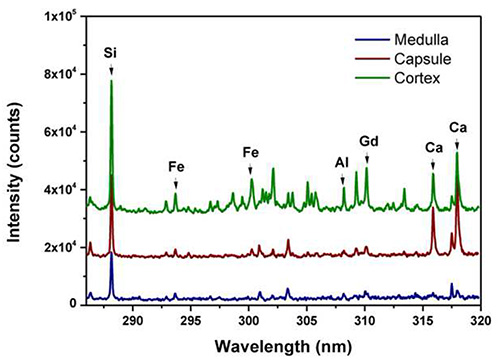
Figure 2. LIBS single-shot spectra obtained in three different regions of the kidney tissue. These regions are the medulla (central part of the kidney), the capsule in red and the cortex in green. Spectra have been shifted vertically for clarity.
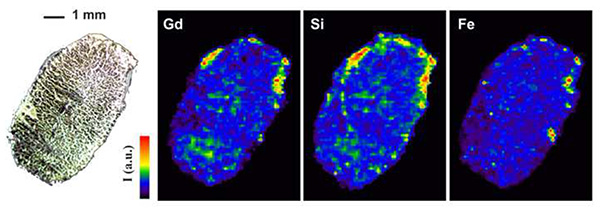
Figure 3. LIBS elemental mapping of Gd, Si, and Fe in a slice of mouse kidney prepared according to the conditions specified in the text. The scale intensity of chemical images is expressed in an arbitrary unit. A natural light photography is also shown in the upper left of the figure.
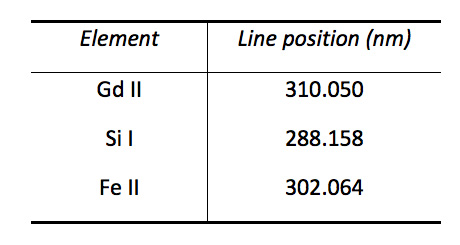
Table 2. Spectral lines used for the detection of Gd, Si, and Fe in a mouse kidney (atomic lines are labeled I and ionic lines are labeled II).
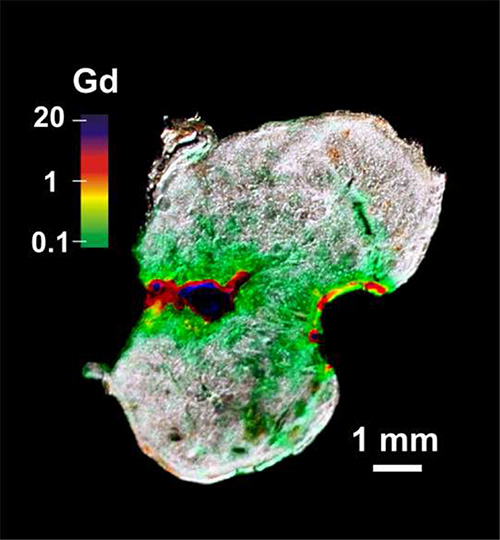
Figure 4. LIBS quantitative mapping of Gd inside of a slice of tumor tissue. The false color scale runs from 0.1 mM (green) to 20 mM (violet).
Discussion
Applied to biological sample, this technique allows the chemical imaging, i.e. the mapping and quantification, of Gd and Si from injected Gd-based nanoparticles in different organs. From the main critical settings, the control of laser properties (wavelength, pulse energy, focusing, and stability) is critical for a precise and fine tissue ablation (i.e. mapping resolution) as well as for sensitivity. Working at high energy provides a better sensitivity but unfortunately generates degraded spatial resolution. Besides, the type of spectrometer used must be checked carefully. Basically, a broadband investigation (using for example an echelle spectrometer) will allow a large choice of elemental species but with low sensitivity, while using a Czerny-Turner spectrometer (equipped with an ICCD or a PMT) will allow detecting fewer elements but with much more sensitivity. All the chosen settings of the probed wavelength range should be adjusted to the purpose of the investigation.
Concerning the biological sample itself, its thickness and hardness might also interfere with the quality of the spectrum. A very thin sample would be totally sprayed and the sample support would be attacked also by the laser shot, whereas a too thick sample might suffer from its in-homogeneity of its constitution that will be reverberated on the in-homogeneity of the amount of sprayed matter (presence of large vessels etc.). From the elemental study, the choice of the support might be anticipated; for example, if the investigated element is Si, the sample should be filed on pure plastic but glass should be avoid as it contains large amounts of Si. Similarly, the sample might be prepared and fixed in specific conditions to prevent any component of the fixative to contaminate the level of the studied element.
This technique might be very useful for the analysis of pre-clinical samples. LIBS could allow the detection of abnormal metallic elements such as Fe and Cu, in particular into brain samples. A particular limitation with Fe should be the high content of Fe in hemoglobin; the sample prepared carefully and the assessment of a calibration assay is mandatory. In the field of biotechnologies applied to biology or medicine, LIBS could allow the detection of any compound or particle that contains a specific element such as Au, Gd, Cu, etc.
The bench-top instrumentation is fully compatible with the standard optical microscopy, which shows its large potential use in Biology and Medicine as a tool for complementary observation of trace metallic elements. This type of elemental imaging, combining high lateral resolution (<50 µm) with low detection limits (in the range of mg/kg), generally involves high level of required equipment, such as synchrotron radiation microanalysis (SXRF) or laser ablation inductively coupled mass spectrometry (LA-ICP-MS), making it very restrictive and incompatible with commercially available microscopic imaging systems. The LIBS instrumentation is not very expensive, depending of the resolution and sensitivity required (price between 100 and 200 k€). Currently a 1 cm2 sample could be analyzed within 30-40 min; this acquisition time might not change after the addition of other microscopic techniques such as fluorescence detection and colored imaging of the sample. The current 100 µm spatial resolution already shows the efficacy for mapping of biological tissues. However, this resolution could be greatly improved by using an optical microscope instead of a simple lens to focus the ablation laser pulse. Micrometer resolution can theoretically be reached, even though the sensitivity will drop, as less material will be ablated. Increasing the resolution to 10 µm would allow an accurate localization of nanoparticles to be observed at the cell level.
To conclude, LIBS is a new technology applicable to the detection and quantification of elements in tissue samples. This approach is sensitive (µg/g), fast (currently 10 measurements/sec), affordable, easy to implement and to use, and fully compatible with conventional optical microscopy.
Declarações
The authors have nothing to disclose.
Acknowledgements
The authors gratefully acknowledge financial support by the Labex-Imust.
Materials
| Laser nanosecond Nd:YAG | Quantel | Brillant | 5ns pulse witdh, wavelength 1064 nm |
| Spectrometer | Andor Technology | Shamrock 303 | with 1200 l/mm blazed at 300 nm grating |
| Detector ICCD | Andor Technology | Istar | 2 ns temporal resolution |
| LIBS Unit | ILM | Homemade Instrumentation | |
| Gd-based nanoparticles | Nano-H | particles | |
| HEPES | Sigma-Aldrich | H4034 | for particle's dilution |
| CaCl2 | Sigma-Aldrich | 21108 | for particle's dilution |
| NaCl | Sigma-Aldrich | S5886 | for particle's dilution |
| mice | Charles River | depending of animal breeding | |
| isoflurane | Coveto / Virbac | for anaesthesia – Isofluranum | |
| isopentane | Sigma-Aldrich | 59060 | to froze the sample slowly |
| liquide nitrogen | Air Liquide | to cool down the isopentane | |
| cryostat | Leica | CM-3050S | to slide the samples |
| petri dishes | Dutscher | 353004 | to stick the sample |
Referências
- Smith, M. A., Harris, P. L., Sayre, L. M., Perry, G. Iron accumulation in Alzheimer disease is a source of redox-generated free radicals. Proceedings of the National Academy of Sciences of the United States of America. 94, 9866-9868 (1997).
- Wang, Y., Zhu, S., Weisman, G. A., Gitlin, J. D., Petris, M. J. Conditional knockout of the Menkes disease copper transporter demonstrates its critical role in embryogenesis. PloS one. 7, (2012).
- Reske-Nielson, E., Lou, H. O., Andersen, P., Vagn-Hansen, P. Brain-copper concentration in Menkes’ disease. Lancet. 1, 613 (1973).
- Hayashi, H., et al. Various copper and iron overload patterns in the livers of patients with Wilson disease and idiopathic copper toxicosis. Medical molecular morphology. 46, (2013).
- Lobinski, R., Moulin, C., Ortega, R. Imaging and speciation of trace elements in biological environment. Biochimie. 88, 1591-1604 (2006).
- Devès, G., Bouhacina, T., Ortega, R. STIM mass measurements for quantitative trace element analysis within biological samples and validation using AFM thickness measurements. Spectrochimica Acta B. 59, 1733-1738 (2004).
- Twining, B. S., et al. Quantifying trace elements in individual aquatic protist cells with a synchrotron X-ray fluorescence microprobe. Analytical chemistry. 75, 3806-3816 (2003).
- Guerquin-Kern, J. L., Wu, T. D., Quintana, C., Croisy, A. Progress in analytical imaging of the cell by dynamic secondary ion mass spectrometry (SIMS microscopy). Biochimica et biophysica acta. 1724, 228-238 (2005).
- Binet, M. R., Ma, R., McLeod, C. W., Poole, R. K. Detection and characterization of zinc- and cadmium-binding proteins in Escherichia coli by gel electrophoresis and laser ablation-inductively coupled plasma-mass spectrometry. Analytical biochemistry. 318, 30-38 (2003).
- Becker, J., Gorbunoff, A., Zoriy, M., Izmer, A., Kayser, M. Evidence of near-field laser ablation inductively coupled plasma mass spectrometry (NF-LA-ICP-MS) at nanometre scale for elemental and isotopic analysis on gels and biological samples. J. Anal. Atom. Spectrom. 21, 19-25 (2006).
- Seeley, E. H., Caprioli, R. M. 3D imaging by mass spectrometry: a new frontier. Analytical chemistry. 84, 2105-2110 (2012).
- Pornwilard, M. -. M., Weiskirchen, R., Gassler, N., Bosserhoff, A. K., Becker, J. S. Novel bioimaging techniques of metals by laser ablation inductively coupled plasma mass spectrometry for diagnosis of fibrotic and cirrhotic liver disorders. PloS one. 8, (2013).
- Cremers, D. A., Radziemski, L. J. . Handbook of laser-induced breakdown spectroscopy. , (2006).
- Miziolek, A. W., Palleschi, V. . Laser-Induced Breakdown Spectroscopy: Fundamentals and Applications. , (2006).
- Motto-Ros, V., et al. Mapping nanoparticles injected into a biological tissue using laser-induced breakdown spectroscopy. Spectrochimica Acta Part B. , (2013).
- Motto-Ros, V., et al. Mapping of native inorganic elements and injected nanoparticles in a biological organ with laser-induced plasma. Applied Physics Letters. 101, (2012).
- Mignot, A., et al. A top-down synthesis route to ultrasmall multifunctional Gd-based silica nanoparticles for theranostic applications. Química. 19, 6122-6136 (2013).
- Lux, F., et al. Ultrasmall rigid particles as multimodal probes for medical applications. Angew Chem Int Ed Engl. 50, 12299-12303 (2011).

