Detection and Removal of Nuclease Contamination During Purification of Recombinant Prototype Foamy Virus Integrase
Summary
Recombinant prototype foamy virus integrase protein is often contaminated with a bacterial nuclease during purification. This method identifies nuclease contamination and removes it from the final preparation of the enzyme.
Abstract
The integrase (IN) protein of the retrovirus prototype foamy virus (PFV) is a model enzyme for studying the mechanism of retroviral integration. Compared to IN from other retroviruses, PFV IN is more soluble and more amenable to experimental manipulation. Additionally, it is sensitive to clinically relevant human immunodeficiency virus (HIV-1) IN inhibitors, suggesting that the catalytic mechanism of PFV IN is similar to that of HIV-1 IN. IN catalyzes the covalent joining of viral complementary DNA (cDNA) to target DNA in a process called strand transfer. This strand transfer reaction introduces nicks to the target DNA. Analysis of integration reaction products can be confounded by the presence of nucleases that similarly nick DNA. A bacterial nuclease has been shown to co-purify with recombinant PFV IN expressed in Escherichia coli (E. coli). Here we describe a method to isolate PFV IN from the contaminating nuclease by heparin affinity chromatography. Fractions are easily screened for nuclease contamination with a supercoiled plasmid and agarose gel electrophoresis. PFV IN and the contaminating nuclease display alternative affinities for heparin sepharose allowing a nuclease-free preparation of recombinant PFV IN suitable for bulk biochemical or single molecule analysis of integration.
Introduction
Biochemical and single molecule studies of protein interactions with DNA require exceptionally pure recombinant proteins. Contaminating nucleases from bacteria can obscure the results of these assays. A contaminating nuclease has been found in preparations of recombinant proteins oxygen scavenger protocatechuate-3,4-dioxygenase (PCD) and prototype foamy virus (PFV) integrase (IN) isolated from Escherichia coli (E. coli)1,2,3.
Retroviral integration assays rely on the conversion of supercoiled DNA to nicked or linear products as a measure of IN activity4. During cellular infection IN joins the two ends of a viral cDNA to the host chromatin5. Each end joining reaction is termed strand transfer. Assays of recombinant IN activity may join two DNA oligomers mimicking the viral cDNA ends to a target DNA in a concerted integration reaction4,5,6,7,8. Alternatively recombinant IN may join only one DNA end in a non-physiologically relevant half site integration reaction9,10. When supercoiled plasmid DNA is the target of integration, concerted integration products are linearized DNA and half site integration products are relaxed circles. These reaction products are identified by their relative mobility during agarose gel electrophoresis1. If the recombinant IN has a contaminating nuclease, there will be spurious relaxed circles or a possibly linearized plasmid confusing the experimental results. Viral DNA oligomers may be fluorescently labeled to conclusively identify integration products, as opposed to nuclease products. However, IN greatly favors supercoiled DNA targets; any loss of supercoiled plasmid to relaxed circles or linear DNA by contaminating nuclease could skew results and interpretation of data11. Thus it is imperative to remove bacterial nucleases from retroviral IN preparations.
PFV IN has a different affinity for heparin sepharose compared to the bacterial nuclease1. PFV IN and the nuclease may be separated by a linear gradient elution from heparin sepharose. The nuclease is not readily detected by an ultraviolet (UV) absorbance at 280 nm peak or by analytical sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Instead, the nuclease is detected by a nuclease activity assay employing the conversion of a supercoiled plasmid to relaxed circles or linear products. Each fraction following heparin sepharose chromatography is tested for nuclease activity. PFV IN and the nuclease contaminant have no difference in affinity for Mono-Q anion resin. There is a small difference in affinity for Mono-S cation resin. However, the Mono-S resolution of bacterial nuclease and PFV IN would not allow efficient separation of the proteins. Ultimately, heparin sepharose affinity purification offers the best separation of bacterial nuclease from PFV IN and has the advantage of unlimited load volume.
Testing for contaminating nuclease activity may be adapted to other proteins. The protein of interest will likely have alternative affinity characteristics than PFV IN; the difference in binding characteristics of the protein of interest and the nuclease contaminant must be empirically determined. This methodology for identifying nuclease contamination may be adapted to other resins including Mono-S cation or Mono-Q anion exchange resins. Affinity and ion exchange resins may offer a reliable method to isolate a recombinant protein of interest from contaminating nucleases with no limits on the volume of protein during chromatography.
Protocol
1. Induce PFV IN Expression in E. coli
- Add 1 μL of PFV IN expression plasmid (10 ng/μL) to 20 μL of E. coli BL21(DE3) pLysS (20 μL aliquot of commercially available cells has >2 x 106 CFU/μg plasmid) in a 1.5 mL tube. Mix gently by finger tapping or flicking the tube. Incubate on ice 5 min.
- Heat shock for exactly 30 s in a 42 ˚C water bath and then immediately return to ice for 2 min.
- Add 80 μL of room temperature super optimal broth with catabolite repression (SOC) media. Incubate for 1 h at 37 ˚C with 225 revolutions per minute (rpm) shaking.
- Plate 25 μL of the mixture on a 100 mm x 15 mm Petri dish containing 25 mL of Luria broth (LB) agar (1 L LB agar: 10 g bacto-tryptone, 10 g NaCl, 5 g yeast extract, 15 g agar) and 100 μg/mL ampicillin (100 mg/mL stock solution).
- Plate the remaining 75 μL of the transformed cells on a second identical plate. Incubate the plates with the lids down overnight at 37 °C.
- Retrieve the plates of transformed E. coli. Use a single colony to inoculate 3 mL of LB (1 L LB: 10 g of bacto-tryptone, 10 g of NaCl, 5 g of yeast extract) supplemented with 100 μg/mL ampicillin in a 14-mL round bottom polypropylene culture tube. Incubate for ~8 h at 37 ˚C with 225 rpm shaking.
- Add the 3 mL of culture to 50 mL LB supplemented with 100 μg/mL ampicillin and 34 μg/mL chloramphenicol (34 mg/mL stock solution) in a 250 mL Erlenmeyer flask. Incubate the culture overnight at 37 °C with 225 rpm shaking.
- Transfer the 53 mL of culture to 1 L LB, 100 μg/mL ampicillin, and 34 μg/mL chloramphenicol in a 4 L Erlenmeyer flask. Incubate at 37 °C with 225 rpm shaking.
- Measure the optical density at 600 nm (OD600) of the culture periodically. Initially, check the culture OD600 every h. As the culture reaches the desired OD600, check every 15 min to not overgrow. Grow the culture to an OD600 between 0.9 and 1.0. Transfer the flask to an ice bath and swirl to reduce the temperature of the culture.
- Transfer 1 mL of the culture to a 1.5 mL tube for later analysis by denaturing SDS-PAGE analysis.
- Pellet the cells in the 1.5 mL tube by centrifugation for 1 min in a microfuge at 14,000 x g at room temperature. Discard the supernatant. Resuspend the pellet in 150 μL phosphate buffered saline (PBS) by pipetting. The PBS may be either with or without CaCl2 and MgCl2.
- Add 150 μL of 2x SDS-PAGE sample buffer (150 mM Tris-HCl, pH 6.8, 1.2% SDS, 30% glycerol, 15% β-mercaptoethanol (βME), 0.0018% bromophenol blue) and mix thoroughly by pipet or vortex. Boil for 3 min. Store at -20 °C.
- Add to the bacterial culture: a final concentration of 0.25 mM isopropyl-beta-D-thiogalactopyranoside (IPTG, 0.25 M stock solution) and 50 μM ZnCl2 (50 mM stock solution). IPTG induces expression of the PFV IN gene from the T7 promoter and ZnCl2 is added as a supplement for the correct folding of an amino terminal zinc finger domain in PFV IN. Incubate the culture at 25 °C with shaking at 225 rpm for 4 h. Immediately transfer the flask to an ice bath. Save 1 mL of culture for SDS-PAGE analysis following the same protocol as above in 1.3.
- Transfer the bacterial culture to appropriately sized centrifuge bottles. For example, a 1 L culture may be transferred to four sterile disposable 250 mL conical bottom polypropylene bottles. Spin the bottles in a refrigerated tabletop centrifuge at 3000 x g for 20 min at 4 ˚C. Discard supernatants by pouring.
- Resuspend all of the pellets from a 1 L bacterial culture in a total volume of 25 mL (6.25 mL per bottle) cold PBS (with or without CaCl2 and MgCl2) by pipetting. Combine and transfer the bacterial suspensions to one 50 mL conical tube. Spin in a refrigerated centrifuge at 3,000 x g for 20 min at 4 °C. Discard the supernatant by pouring or pipetting.
- Resuspend the bacterial pellet in 10 mL resuspension buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, and 500 μM phenylmethylsulfanoxide (PMSF) protease inhibitor) by pipetting. Snap freeze the pellet by placing the tube in a Dewar flask with liquid nitrogen sufficient to submerge the 50 mL tube. Carefully remove the tube from liquid nitrogen and store at -80 °C.
Note: Bacterial pellets may be stored for several months at -80 °C. We have seen no loss of active protein following -80 °C storage for two years. - Analyze the pre-induction and post-induction samples by 8.3 cm wide, 7.3 cm high, 0.75 mm thick SDS-PAGE gels with 1.5 mL 6% polyacrylamide stacking layer (125 mM Tris-HCl, pH 6.8, 6% acrylamide, 0.1% SDS, 0.1% ammonium persulfate, 0.001% TEMED) and 3.5 mL 10% polyacrylamide resolving layer (375 mM Tris-HCl, pH 8.8, 10% acrylamide, 0.1% SDS, 0.1% ammonium persulfate, 0.001% TEMED)12.
- After pouring the stacking gel, insert a 10 well comb. When the gel has polymerized, assemble the PAGE apparatus, and add sufficient SDS-PAGE running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS).
- Load 10 μL of each sample and 4 μL of prestained protein standards. Run at 16.5 V/cm until the dye front reaches the bottom of the gel, approximately 60 min.
- Disassemble the gel plates and transfer the gel to a plastic tub. Add Coomassie Brilliant Blue staining solution (0.1% Coomassie brilliant blue R-250, 40% methanol, 10% acetic acid) to generously cover the gel and stain for 20 min at ambient temperature.
- Remove the staining solution by pouring and replace with destain solution (40% methanol, 10% acetic acid) until bands are readily visible. Remove the destaining solution by pouring and replace with deionized water. PFV IN should be easily identified in the post-induction sample. PFV IN with a hexahistidine tag has a molecular weight of 47374 Da.
- If PFV IN is not visible, repeat the induction starting with a different colony from one of the transformation plates.
2. Nickel Affinity Chromatography
- Thaw one bacterial pellet on ice.
Note: This will take significant time (approximately 2 – 3 h). When the pellet has thawed to a cell slurry, add an additional 500 μM PMSF (100 mM stock solution). - Keep the 50 mL tube of bacteria slurry on ice. Sonicate the bacteria at 30% amplitude for 30 s (1 s on, 1 s off) per pellet derived from 1 L culture using a sonicator with a 0.5 inch diameter bio horn with a 0.125 inch diameter tapered microtip, a frequency of 20 kHz and maximum power of 400 W.
- Transfer the cell sample to a cold ultracentrifuge tube. Use polycarbonate bottle assemblies (25 mm diameter, 89 mm height) compatible with a Type 60 Ti fixed angle rotor. Alternative tubes and rotors may be used if the same gravitation force is achieved. Spin 120,000 x g for 60 min at 4 °C. There should be an obvious pellet. The supernatant may have a yellow color.
- Transfer the supernatant by pouring or pipetting to a cold 50 mL conical tube. Note the volume; this volume should be approximately the volume of resuspension buffer used prior to snap freezing. If the supernatant is viscous, it may be contaminated by bacterial DNA which could clog the chromatography column. In this case, repeat the ultracentrifugation to pellet the bacterial DNA.
- Prepare 500 mL Buffer A (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 500 μM PMSF) and 500 mL Buffer B (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 500 μM PMSF, 200 mM imidazole).
Note: In this protocol, keep the fast protein liquid chromatography (FPLC) system, all buffers and the sample at 4 °C in a cold room.- Sterile filter Buffers A and B with 0.2 μm disposable filter units. Depending on the FPLC system, wash pump A and pump B with Buffer A and Buffer B, respectively. Flow 90% Buffer A and 10% Buffer B through the system until the conductivity and UV readings stabilize. The maximum flow rate will depend on the instrument; use a flow rate of 5 mL/min with a maximum pressure limit of 1.0 MPa.
- Prepare a 110 mm long, 5 mm diameter FPLC column with 1.5 mL nickel-charged resin (maximum binding capacity 50 mg/mL, maximum pressure 1 MPa). The column may be prepared the day before purification and stored at 4 ˚C.
- Connect the column to the FPLC instrument. Equilibrate the column with 20 mL of 90% Buffer A and 10% Buffer B (20 mM imidazole) at a flow rate of 0.5 mL/min with a maximum pressure limit of 0.5 MPa. The instrument should be collecting real time data of conductivity and UV absorbance at 280 nm (A280). The conductivity and UV readings should stabilize. If these readings have not stabilized, continue to flow buffers through the column until they are stable.
- Load the protein sample (~10 mL per pellet) to the column at a flow rate of 0.15 mL/min with a maximum pressure limit of 0.5 MPa. The flow rate and column pressure remains the same throughout the purification. Collect the flow through in a 50 mL tube.
- Wash the column with 20 mL 90% Buffer A and 10% Buffer B. Collect the wash in a 50 mL tube. Elute the proteins with a 20 mL linear gradient of 10% to 100% Buffer B (20 mM to 200 mM imidazole).
- Finally, wash the column with 5 mL 100% Buffer B (200 mM imidazole). Collect the gradient and final wash in 0.27 mL fractions.
- Combine 15 μL of 2x SDS-PAGE sample buffer with 15 μL of the flow through, wash, and peak A280 fractions. Boil the samples for 3 min.
- Prepare two 10% SDS-PAGE gels as described previously in step 1.6 except with 15 well combs. Load 10 μL of each sample to the gels and 4 μL of prestained protein standards. Run, stain, and analyze the gels as described in step 1.6.
- Combine fractions that appear to contain nearly pure PFV IN based on visual inspection. Measure the volume by pipetting, typically 8 – 10 mL.
- Using a spectrophotometer, measure the A280 of the combined fractions and calculate the total amount of PFV IN protein; our typical yield is ~10 mg per L of induced culture. The extinction coefficient of PFV IN with the hexahistidine tag is 59360 cm-1 M-1.
- To remove the hexahistidine tag, supplement the PFV IN with 10 mM dithiothreitol (DTT, 1 M stock solution) and 0.1 mM EDTA, pH 8.0 (0.5 M stock solution). Add 15 μg of human rhinovirus (HRV) 3C protease per mg PFV IN. Incubate at 4 ˚C overnight. Cleavage reduces the PFV IN molecular weight from 47374 Da to 44394 Da and may be verified by 10% SDS-PAGE analysis.
3. Heparin affinity chromatography
- Prepare 250 mL Buffer C (50 mM Tris-HCl, pH 7.5, 10 mM DTT, 0.1 mM EDTA, 500 μM PMSF) and 250 mL Buffer D (50 mM Tris-HCl, pH 7.5, 10 mM DTT, 0.1 mM EDTA, 500 μM PMSF, 1 M NaCl).
Note: In this protocol, the FPLC system, buffers, and sample are kept in a cold room at 4 ˚C.- Sterile filter Buffers C and D with 0.2 μm disposable filter units. Depending on the FPLC system, wash pump A and pump B with Buffer C and Buffer D, respectively. Flow 80% Buffer C and 20% Buffer D through the system until the conductivity and UV readings stabilize. The maximum flow rate will depend on the instrument; we routinely use a flow rate of 5 mL/min with a maximum pressure limit of 1.0 MPa.
- Prepare an 80 mm long, 5 mm diameter FPLC column with 1 mL of heparin sepharose resin (maximum binding capacity 2.0 mg of bovine antithrombin III per mL resin; maximum pressure 1.4 MPa). The column may be prepared the day before purification and stored at 4 °C. Connect the column to the FPLC instrument.
- Equilibrate the column with 20 mL of 80% Buffer C and 20% Buffer D (200 mM NaCl) at a flow rate of 0.5 mL/min with a maximum pressure limit of 1 MPa. The instrument should be collecting real time data of conductivity and UV absorbance at 280 nm (A280). The conductivity and UV readings should stabilize. If these readings have not stabilized, continue to flow buffers through the column until they are stable.
- Reduce the NaCl concentration of the PFV IN sample to 200 mM by adding 1.5 volumes Buffer C. For example, add 15 mL of Buffer C to 10 mL PFV IN. Our total volume is typically ~25 mL.
- Load the diluted PFV IN sample to the heparin sepharose column at 0.5 mL/min flow rate with maximum pressure limit 1.0 MPa. The flow rate and column pressure remains the same throughout the purification. Collect the flow through in a 50 mL conical tube.
- Wash the column with 20 mL of 80% Buffer C and 20% Buffer D (200 mM NaCl), collecting the wash in a 50 mL conical tube. Elute the column with a 30 mL linear gradient of 20% to 100% Buffer D (200 mM to 1 M NaCl).
- Wash the column with 5 mL of 100% Buffer D. Collect the gradient and final wash in 0.37 mL fractions.
- Analyze load, flow through, wash, and fractions by 8% SDS-PAGE as described in 2.9. PFV IN following cleavage of the hexahistidine tag has a molecular weight of 44394 Da and the extinction coefficient remains 59360 cm-1 M-1 without the hexahistidine tag. Store all fractions at 4 ˚C until the completion of a nuclease assay.
4. Nuclease assay
- Identify heparin sepharose fractions that appear to contain nearly pure PFV IN. Combine 3 μL of heparin fraction and 50 ng of 3 kb supercoiled plasmid DNA in reaction buffer (10 mM HEPES, pH 7.5, 110 mM NaCl, 5 mM MgSO4, 4 μM ZnCl2, 10 mM DTT) with a final volume of 15 μL. Incubate at 37 ˚C for 90 min. Include a negative control with no PFV IN.
- Stop the reaction with 1 mg/mL proteinase K (20 mg/mL stock solution) and 0.5% SDS (10% stock solution). Incubate at 37 °C for 1 h. Samples may be stored at -20 °C for later analysis.
- Prepare a 125 mL gel of 1% weight per volume (w/v) agarose in 1x TAE buffer (40 mM Tris-acetate, 1 mM EDTA) with 0.1 µg/mL ethidium bromide (10 mg/mL stock solution)13. Melt the agarose solution and pour to a 15 cm x 10 cm gel casting tray. Insert a 15 well comb with 5 mm wide, 0.75 mm thick wells. Thin wells yield better band resolution compared to 1.5 mm thick wells. Allow the gel to solidify at ambient temperature.
- Immerse the gel in 1x TAE with 0.1 µg/mL ethidium bromide. Add 3.5 μL of 6x loading dye (50% glycerol, 0.15% Orange G dye) to the nuclease assay reactions. Load the entire volume to the gel. Run the gel at constant voltage, 10 volts per cm (100 V) at ambient temperature for 1 h or until the Orange G dye front reaches the end of the gel.
- Immediately image the agarose gel with a fluorescent scanner set to detect ethidium bromide. Linear DNA should run true to size at 3 kb, supercoiled DNA should run faster (~2 kb), and relaxed circle DNA should run slower (~3.5 kb).
- Using image analysis software, calculate the pixel volume of the supercoiled, linear, and relaxed circle plasmid in each lane. Call the sum of the pixel volumes for these three DNA forms the "total DNA" value and use it to calculate the percentage of linear and relaxed circles. For example, the pixel volume of relaxed circles is divided by the total DNA pixel value in that lane, multiply this number by 100 to determine a percentage. Fractions that contain contaminating nuclease display a higher percentage of linear and relaxed circles compared to the negative control.
- Combine heparin sepharose fractions that do not display contaminating nuclease activity. Dialyze in 10 mm 6 – 8 kDa molecular weight cutoff (MWCO) dialysis tubing at 4 °C overnight against 1 L of 50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 50 μM ZnCl2, 10 mM DTT, 20% glycerol.
- Recover the nuclease-free PFV IN sample from dialysis tubing. Measure the A280 and calculate the final protein concentration. Aliquot and snap freeze in liquid nitrogen. Store aliquots at -80 °C.
Representative Results
Recombinant PFV IN is often contaminated with a bacterial nuclease1. Biochemical integration assays depend on the quantitation of the conversion of supercoiled plasmid DNA to relaxed circles and linear products. The presence of a contaminating nuclease could lead to spurious quantitation of these assays. Expression of PFV IN with a hexahistidine tag is induced in E. coli (Figure 1) and first purified by nickel affinity chromatography (Figure 2). The fractions with nearly pure PFV IN are combined and the hexahistidine tag is cleaved by a protease. We have determined that PFV IN and the contaminating nuclease have different affinities for heparin sepharose chromatography (Figure 3)1. PFV IN fractions following heparin sepharose chromatography are incubated with a supercoiled plasmid to assay for relaxed circles and linearized plasmid, the products of nuclease activity (Figure 4). This assay allows the identification of protein fractions without the nuclease (Figure 5). These fractions may be combined, dialyzed, and frozen for future experiments.
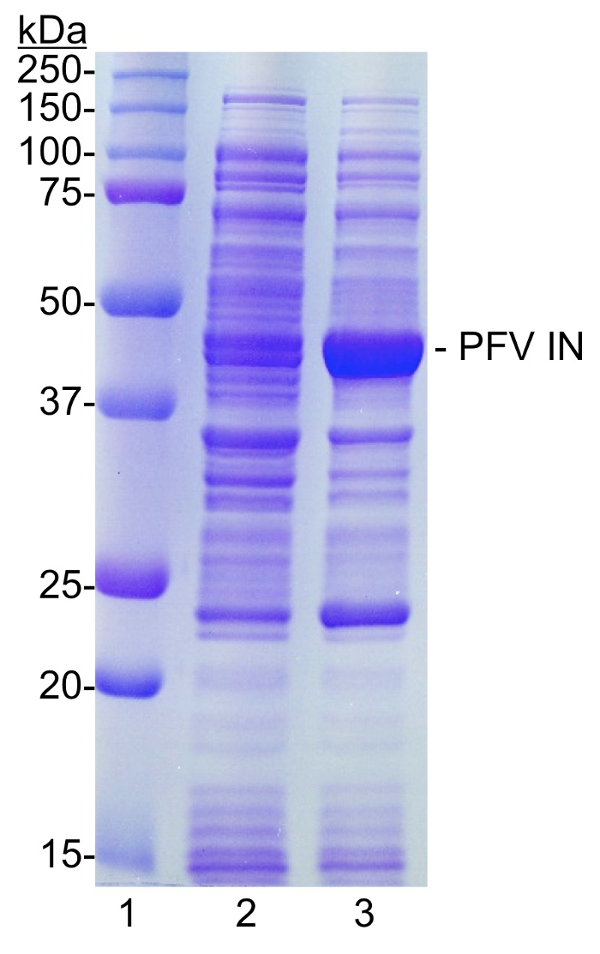
Figure 1: Induction of PFV IN in E. coli . Cells from cultures before (lane 2) and after (lane 3) induction are analyzed for the presence of PFV IN, 47374 Da. Samples were separated by 10% SDS-PAGE and stained with Coomassie brilliant blue. Lane 1, molecular weight markers (kDa). Please click here to view a larger version of this figure.
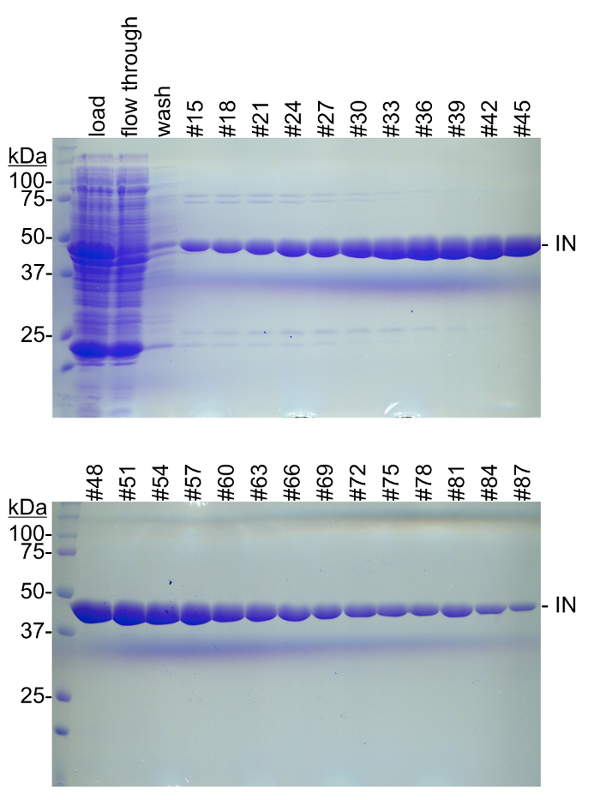
Figure 2: Nickel affinity chromatography fractions. Soluble bacterial lysates were separated by nickel affinity chromatography. PFV IN with a hexahistidine tag was eluted with a linear gradient of 20 mM to 200 mM imidazole. Fractions were evaluated by 10% SDS-PAGE and stained with Coomassie brilliant blue. PFV IN, 47374 Da, is readily apparent in the sample loaded to the column (load), but is less apparent in the flow through or wash. Fractions that elute early in the imidazole gradient commonly display a slower mobility contaminant near 75 kDa as well as some faster mobility contaminants at or below 25 kDa. At higher concentrations of imidazole there are no readily apparent contaminants and PFV IN appears to be relatively pure. In this example, fractions #33 through #84 were combined and further purified. Molecular weight markers (kDa) are on the left of each gel. Please click here to view a larger version of this figure.
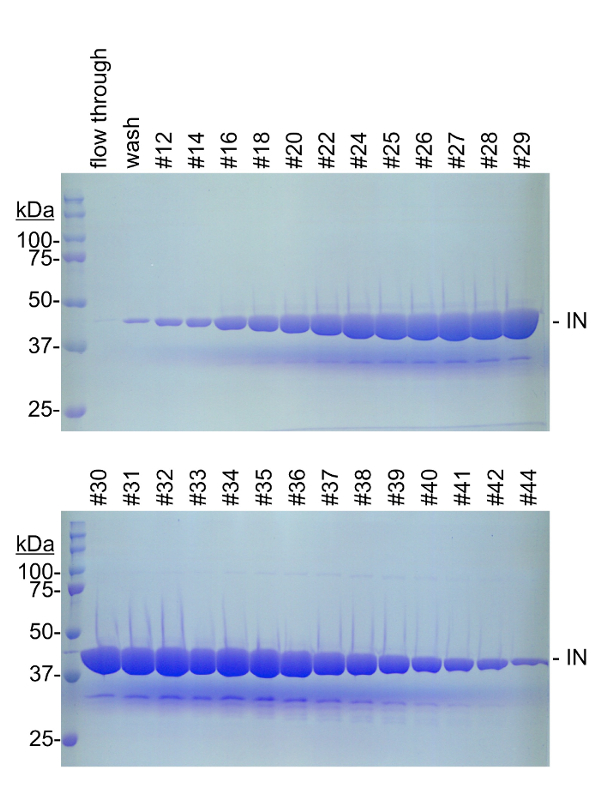
Figure 3: Heparin sepharose chromatography of PFV IN 1 . Following nickel affinity and protease removal of the hexahistidine tag, PFV IN (44394 Da) was fractionated by heparin sepharose. Proteins were eluted with a linear gradient from 200 mM to 1 M NaCl. Even fractions #12 to #44 were evaluated by 8% SDS-PAGE stained with Coomassie blue. Molecular weight markers (kDa) are on the left of each gel. These fractions were tested for nuclease activity. Please click here to view a larger version of this figure.
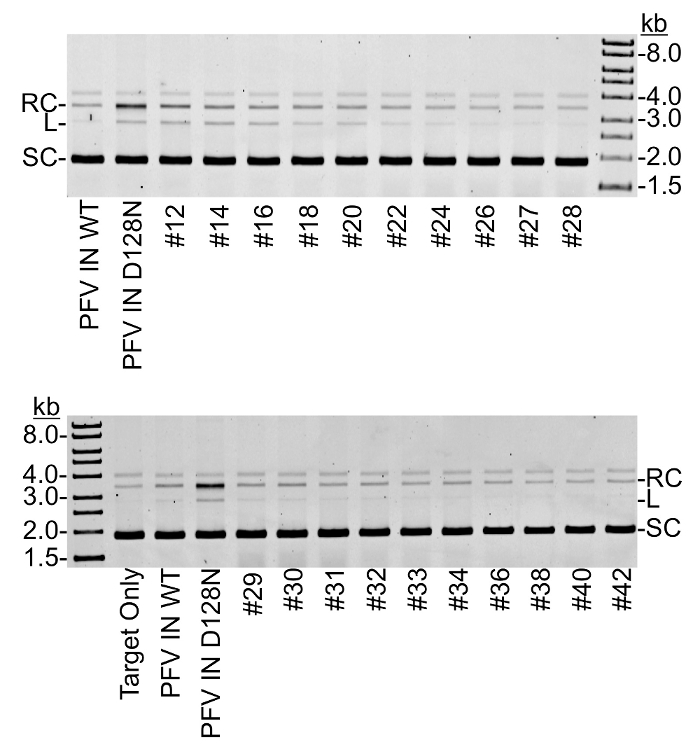
Figure 4: Nuclease assay of heparin sepharose fractions1 . Each heparin sepharose fraction was added to supercoiled plasmid DNA. Fraction numbers correlate to those shown in Figure 3. Following incubation, the nuclease reactions were deproteinated and separated by 1% agarose with ethidium bromide. Negative controls include plasmid DNA with no protein (Target Only) and a previous purification of wild type PFV IN known to be free of nuclease (PFV IN WT). A positive control for contaminating nuclease activity is a previous purification of catalytically inactive PFV IN that is known to have contaminating nuclease (PFV IN D128N). DNA size markers are shown in kb. Supercoiled plasmid (SC), linear plasmid (L), and relaxed circles (RC) are indicated. Please click here to view a larger version of this figure.
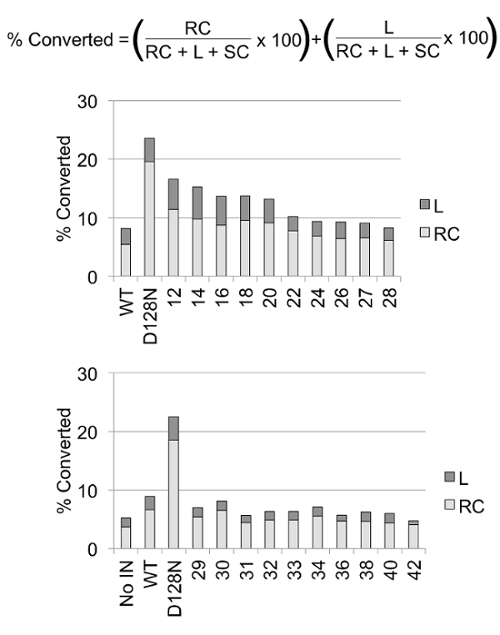
Figure 5: Quantitation of nuclease assays1. Agarose gels were scanned and pixel values were quantified for supercoiled (SC), linear (L), and relaxed circle (RC) bands in each lane. Fraction numbers correlate to those shown in Figures 3 and 4. The percentages of linear and relaxed circles were calculated using the pixel values and the equation shown. For example, the pixel values for the bands of fraction #12 are: 817636 supercoiled plasmid, 50467 linear plasmid, and 112052 relaxed circles. The total DNA pixel value for fraction #12 is the sum of these values, 980155. The percentage of linear DNA is 50467 multiplied by 100 for the percentage and divided by the total DNA value equaling 5.1%. The percentage of relaxed circles is 112052 multiplied by 100 and divided by the total DNA value equaling 11.4%. The sum of linear and relaxed circle percentages is 16.5%. Negative control plasmid DNA with no protein (No IN, bottom graph) indicates that this preparation of plasmid included 4.4% of the total DNA as linear or relaxed circles. A second negative control was a previous purification of wild type PFV IN (WT) that displayed 8.6% of the total DNA as linear or relaxed circles. These two control lanes indicate the background level of nicked or linearized plasmid present with the substrate alone and with PFV IN. A positive control is a previous purification of catalytically inactive PFV IN (D128N) which had 23% of the total DNA as linear and relaxed circles. The DNA exposed to heparin sepharose fractions #12 through #22 resulted in >10% of the DNA as linear and relaxed circles. In this example, fractions #24 to #42 displayed <10% conversion to linear and relaxed circles. These fractions were combined and dialyzed. Aliquots were frozen and stored at -80 °C for future use. Please click here to view a larger version of this figure.
Discussion
Recombinant proteins that interact with DNA, such as DNA repair proteins, oxygen scavengers for single molecule microscopy applications, or retroviral integrases, should be free of contaminating bacterial nucleases2,3. These contaminants may confuse the interpretation of results during bulk biochemical or single molecule assays.
We have found that a bacterial nuclease frequently co-purifies with PFV IN. However, PFV IN displays an affinity for heparin sepharose that is readily distinguishable from the bacterial nuclease contaminant1. The key to preparation of nuclease-free PFV IN is the careful quantitation of nuclease activity in each fraction following gradient elution from heparin sepharose. The gradient must be sufficiently shallow to allow the separation of PFV IN from the nuclease contaminant; a steep gradient would not be compatible with efficient separation. In addition, the plasmid DNA substrate should have a low percentage of relaxed circles to reduce the background of the nuclease assay. If the plasmid DNA substrate has a high background of relaxed circles, a new purification of plasmid DNA should be performed. The nuclease activity assay reveals the presence of the contaminating nuclease and fractions that should be discarded.
In some cases, size exclusion chromatography (SEC) may be an effective method for removal of a contaminating nuclease2. Previous groups have used SEC during purification of PFV IN4. SEC has a limited load volume and tends to reduce the final protein concentration. The major advantage to affinity and ion exchange chromatography is the ability to load a significantly greater volume than SEC.
Testing for nuclease activity may be adapted to different proteins that co-purify with a bacterial nuclease. Here we identify that PFV IN and a contaminating nuclease may be separated by heparin sepharose chromatography. We have previously shown that PFV IN has a similar profile to the nuclease with Mono-S cation and Mono-Q anion exchange chromatography making these resins unsuitable for this protein. Depending on the affinity characteristics of the desired protein in relation to the nuclease, this protocol may be used with affinity or ion exchange resins and gradient elution. The key to effective removal of nuclease contamination from a protein of interest is a difference in affinity for a resin. Each fraction must be assayed for nuclease activity. The chromatography resins of this protocol may not be directly applied to all proteins that co-purify with bacterial nucleases. However, the overall concept of using an affinity or ion exchange resin to separate the nuclease from a protein of interest and assaying fractions for nuclease activity may be broadly applicable.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was supported by NIH AI099854 and AI126742 to KEY.
Materials
| BL21/DE3 Rosetta E. coli | EMD Millipore | 70956-4 | |
| LB broth | EMD biosciences | 1.10285.0500 | |
| Ampicillin | Amresco | 0339 | |
| Chloramphenicol | Amresco | 0230 | |
| PBS | Sigma-Aldrich | D8537 | |
| IPTG | Denville Scientific | CI8280 | |
| ZnCl2 | Sigma-Aldrich | 208086 | |
| Tris Ultra Pure | Gojira Fine Chemicals | UTS1003 | |
| NaCl | P212121 | RP-S23020 | |
| PMSF | Amresco | 0754 | |
| Imidazole | Sigma-Aldrich | I0250 | |
| DTT | P212121 | SV-DTT | |
| UltraPure EDTA | Invitrogen/Gibco | 15575 | |
| MgSO4 | Amresco | 0662 | |
| Agarose | Denville Scientific | CA3510 | |
| Ethidium bromide | Thermo Fisher Scientific | BP1302 | |
| Glycerol | Fisher Scientific | G37-20 | |
| Ni-NTA Superflow | Qiagen | 30430 | |
| Heparin Sepharose 6 Fast Flow | GE Healthcare Life Sciences | 17-0998-01 | |
| HRV14 3C protease | EMD Chemicals | 71493-3 |
Referências
- Lopez, M. A., Mackler, R. M., Yoder, K. E. Removal of nuclease contamination during purification of recombinant prototype foamy virus integrase. J Virol Methods. 235, 134-138 (2016).
- Senavirathne, G., et al. Widespread nuclease contamination in commonly used oxygen-scavenging systems. Nat Methods. 12 (10), 901-902 (2015).
- Gunn, K. H., Marko, J. F., Mondragon, A. An orthogonal single-molecule experiment reveals multiple-attempt dynamics of type IA topoisomerases. Nat Struct Mol Biol. 24 (5), 484-490 (2017).
- Valkov, E., et al. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 37 (1), 243-255 (2009).
- Coffin, J. M., Hughes, S. H., Varmus, H. E. . Retroviruses. , (1997).
- Li, M., Craigie, R. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J Biol Chem. 280 (32), 29334-29339 (2005).
- Li, M., Mizuuchi, M., Burke, T. R., Craigie, R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 25 (6), 1295-1304 (2006).
- Sinha, S., Grandgenett, D. P. Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. J Virol. 79 (13), 8208-8216 (2005).
- Goodarzi, G., Im, G. J., Brackmann, K., Grandgenett, D. Concerted integration of retrovirus-like DNA by human immunodeficiency virus type 1 integrase. J Virol. 69 (10), 6090-6097 (1995).
- Vora, A. C., Grandgenett, D. P. Assembly and catalytic properties of retrovirus integrase-DNA complexes capable of efficiently performing concerted integration. J Virol. 69 (12), 7483-7488 (1995).
- Jones, N. D., et al. Retroviral intasomes search for a target DNA by 1D diffusion which rarely results in integration. Nat Commun. 7, 11409 (2016).
- Lee, P. Y., Costumbrado, J., Hsu, C. Y., Kim, Y. H. Agarose gel electrophoresis for the separation of DNA fragments. J Vis Exp. (62), (2012).

