Detection and Quantification of Plasmodium falciparum in Aqueous Red Blood Cells by Attenuated Total Reflection Infrared Spectroscopy and Multivariate Data Analysis
Summary
Here, we present a protocol for the detection and quantification of Plasmodium falciparum in infected aqueous red blood cells using an attenuated total reflection infrared spectrometer and multivariate data analysis.
Abstract
We demonstrate a method of quantification and detection of parasites in aqueous red blood cells (RBCs) by using a simple benchtop Attenuated Total Reflection Fourier Transform Infrared (ATR-FTIR) spectrometer in conjunction with Multivariate Data Analysis (MVDA). 3D7 P. falciparum were cultured to 10% parasitemia ring stage parasites and used to spike fresh donor isolated RBCs to create a dilution series between 0–1%. 10 µL of each sample were placed onto the center of the ATR diamond window to acquire the spectrum. The sample data was treated to improve the signal to noise ratio and to remove the contribution of water, and then the second derivative was applied to resolve spectral features. The data were then analyzed using two types of MVDA: first Principal Component Analysis (PCA) to determine any outliers and then Partial Least Squares Regression (PLS-R) to build the quantification model.
Introduction
Malaria is among the most devastating diseases of our time; over half the population lives at risk in endemic regions and it disproportionately burdens the poor1,2,3,4. A large part of the issue is the asymptomatic carriers and early stage patients that act as reservoirs for mosquito vectors5, causing spikes of infection during wet seasons and allowing it to persist in communities. Malaria is caused by five Plasmodium parasites, the most deadly of which is P. falciparum which causes the most severe form of the disease2.
Currently, techniques for the diagnosis of malaria are less than perfect. Optical microscopy, the current gold standard, can only detect 62-88 parasites/µL depending on the method used6. Furthermore, due to the intensiveness and high skill required, in many regions microscopy misdiagnoses >50% of cases, especially those with low parasitaemia levels2, which can be directly attributed to the lack of resources in the area and results in the misuse of anti-malarial drugs. The other 2 main diagnostic methods are Rapid Diagnostic Tests (RDTs), which utilize antibodies for the detection, and Polymerase Chain Reaction (PCR) assays, which discriminate and quantify parasites from DNA. Currently, RDTs are only able to detect P. falciparum and P. vivax of at least 100 parasites/µL of blood resulting in rarer forms of the disease being untreated.7,8 In contrast, PCR assays discriminate and quantify different species of Plasmodium at a sensitivity of 0.0004-5 parasites/µL of blood. However, it requires expensive reagents, equipment and technical skill, and thus is not suitable for the field application.
A highly sensitive, reliable and affordable technique is essential to improve diagnosis times, and thus improving patient outcomes, and making disease elimination possible. Attenuated Total Reflection Fourier transform (ATR-FTIR) spectroscopy offers a potential solution to this problem. Previous work has shown that it was possible to detect and quantify P. falciparum in methanol fixed blood films achieving the detection limits of <1 parasite/µL of blood (<0.0002 % parasitemia)9, which is comparable to PCR methods. Recent studies have shown that it is possible to detect and quantify parasites in aqueous samples and thus eliminate the fixation step. However, factors such as water vapor, spectral noise and data treatment need to be taken into consideration for optimal results10.
This protocol aims to show new users how to acquire ATR-FTIR spectra and prepare a regression model for the detection of P. falciparum from aqueous Red Blood Cells (RBC) samples10.
Protocol
Please consult appropriate Material Safety Data Sheets (MSDS) and seek appropriate Biosafety Level 2 (BSL-2) training. All culturing steps must be done in a BSL-2 cabinet using aseptic technique, meaning there is a risk of exposure to harmful UV radiation from decontamination steps, needle stick injuries, and potential biological exposure and infection if the parasite culture enters any injuries. Furthermore, stock blood from blood banks is only screened for certain diseases and the potential for spreading blood borne diseases is a potential risk. Seek immediate medical aid in the occurrence of injury.
1. Preparation and Measurement of 3D7 Plasmodium falciparum Parasite Dilution Series
NOTE: Plasmodium sp. culture is highly sensitive. Use fresh/unexpired reagents and feed the parasites regularly by changing the media. Feed cultures below 5% parasitemia every second day; feed cultures between 6-10% parasitemia once or twice a day; and feed cultures between 11–20% up to 4 times a day. Parasites that are beginning to starve will lose their shape and begin to contract. In such a case, feed and dilute immediately by changing the media and adding stock RBCs. Collect the donor blood in blood collection tubes containing heparin as the anticoagulant and measure within the first 6 h.
- Culture a total of 30 mL of 3D7 strain Plasmodium falciparum parasites according to the standard protocol until the parasitemia reaches 10% rings.
- Synchronization of parasites
- 11 h after shizogeny into the parasite lifecycle, resuspend the culture and transfer into a 50 mL conical tube using an automated pipette.
- Centrifuge the culture at 300–400 x g and standard laboratory conditions (25°C and 100 kPa) for 5 min.
- Remove the supernatant by drawing it up with a pipette or using a Pasteur pipette attached to a vacuum without disturbing the pellet. Discard waste media into a 10% bleach solution.
- Slowly add 12–15 mL of 4% sorbitol solution to the pellet using an automated pipette and mix the culture by capping and inverting the tube until the culture is homogenous.
- Incubate the culture at 37 °C for 15 min.
- Centrifuge the culture at 300–400 x g and standard laboratory conditions for 5 min.
- Remove the supernatant as described in step 1.2.3.
- Add 10–15 mL of 0.9% saline to the pellet and mix the solution by capping and inverting the tube until the culture is homogenous.
- Repeat the steps 1.2.6–1.2.8 twice more to wash all remnants of sorbitol.
- Preparation of the parasitemia dilution series
- Wash stock RBCs as in steps 1.2.6–1.2.9.
- Centrifuge the culture at standard laboratory conditions, stock RBCs at 300–400 x g for 5 min and discard supernatant as in step 1.2.3.
- Label microcentrifuge tubes as 0%, 0.010%, 0.025%, 0.075%, 0.100%, 0.250%, 0.750% and 1.000%, and add 0 µL, 1 µL, 2.5 µL, 7.5 µL, 10 µL, 25 µL, 75 µL and 100 µL of culture respectively using a 0.2-20 µL pipette.
- Add 100 µL, 99 µL, 97.5 µL, 92.5 µL, 80 µL, 75 µL, 25 and 0 µL of stock RBCs to the tubes labelled 0%, 0.010%, 0.025%, 0.075%, 0.100%, 0.250%, 0.750%, and 1.000%, respectively.
- Centrifuge the fresh donor blood collected in anticoagulant tubes at 1,200 x g and standard laboratory conditions for 10 min.
- Remove the plasma by drawing it up with a pipette and discarding as described in step 1.2.3.
- Add 900 µL of isolated RBCs into each microcentrifuge tube and mix thoroughly by inverting 10x.
- Spectral acquisition
- Using the accompanying spectral acquisition program, click "Instrument Set-Up" and set the data collection parameters to the following: 128 scans for background; 32 scans for sample; and resolution to 8/cm over the maximum sample range of the instrument.
- Set the temperature of the ATR-FTIR spectrometer per manufacturer's recommendation or overnight.
- Clean the crystal by gently scrubbing in a circular motion using lint free wipes dampened with ultrapure water. Then thoroughly dry using another lint free wipe.
- Take a background measurement of the air by clicking "Background Measurement". Every 20 min, clean the crystal as in step 1.4.3 and repeat this measurement.
- Open the live view by clicking "Preview". Observe a flat, horizontal baseline that indicates that the crystal is clean.
- Pipette 10 µL of deionized water directly on to the middle of the crystal and click "Measure Sample". Repeat this water measurement after every fifth sample.
- Dry the crystal using a lint free wipe and a gentle circular motion.
- Pipette 10 µL of sample and click "Measure Sample".
- Clean the crystal between samples as in step 1.4.3.
- Repeat until 3 replicates are acquired, making sure to randomize the order of both the samples and the replicates.
2. Multivariate Data Analysis (MVDA)
NOTE: Data treatment must be done over the whole dataset in order to avoid noise and the addition of peaks of non-biological origin. In contrast, analyze over the biologically relevant regions: 2980-2800/cm and 1750-850/cm.
- Data treatment
NOTE: Two software, e.g., Matlab (henceforth referred to as software 1) and The Unscrambler X (henceforth referred to as software 2) are used as examples for MVDA. Software 1 has the capability of performing MVDA without the addition of a graphical user interface (GUI). However, it is recommended to purchase a GUI (Table of Materials). The following instructions when referring to software 1 will assume that is in conjunction with the commercially available GUI toolbox, and the example data treatment script has been attached.- Open the appropriate multivariate data analysis software, click "Import Data" and select the type of file for analysis.
- In software 1, input "Analysis" into the command window to open the GUI. Right click the X box to find "Import Data".
- In software 2, click the tab "File" to find "Import".
- Import sample, water and baseline spectra as data sets into the workspace by selecting all spectra in each set separately and clicking "Open" and give each set a short name, e.g., 'wat' for dataset of water spectra.
- Select new table/matrix by clicking "New Matrix/Vector" and generate a n×1 vector, where n is the number of samples. Input the parasitemia of each sample and give the vector the name 'Parasitemia'.
- In software 1, click "New Variable" in the command window.
- In software 2, click the icon "New Matrix".
- Plot data by clicking the "Plot Data Icon". Inspect the spectra for water vapor effects by clicking "Zoom" and zooming in on 1800-1400/cm; most clearly observed as short, sharp, narrow peaks along the slopes of the amide I and amide II bands.
- In cases of extreme water vapor, open edit/pre-process data tab, and select "Smoothing". Reduce the noise and/or strong water vapor contributions by smoothing the sample and water spectra using up to 25 points of smoothing or use a water vapor correction method.
- Correct non-horizontal baseline by using the baseline correction algorithm if appropriate, under the same edit/pre-process data tab in step 2.1.4.
- Average water spectra and copy the rows into an n×m matrix equivalent to the sample dataset and reduce it to 70% intensity by multiplying it by 0.7.
- In software 1, do so in the command window by inputting the following script "AverageWater=mean(WaterDataset)". Then copy-paste the rows to match the sample data set. To reduce the intensity, input "AverageWater70= AverageWater*0.70".
- Subtract average water spectra from each sample spectrum.
- In software 1, do so in the command window by inputting the following script "WaterCorrectedData=SampleDataset-AverageWater70".
- Open the edit/pre-process data tab to apply a second derivative function, and then normalize the data by selecting single normal variate (SNV) function and mean center data.
- In software 1, do so in one go. First select "Derivative", and input 25 points of smoothing, polynomial order of 3 and derivative order on the sample set using 25 points of smoothing and a Savitzky-Golay function. Then select "SNV" and "Mean Center". Click "Okay/Apply".
- Open the edit/pre-process data tab and in "Column Variables", select 2,980–2,800/cm and 1,750–850/cm by making sure only their boxes are ticked.
- Open the appropriate multivariate data analysis software, click "Import Data" and select the type of file for analysis.
- Data analysis
- Principal Component Analysis (PCA)
- Click "Analysis | Decomposition" and select PCA.
- In software 1, click "Build Model".
- In software 2, input the PCA methods. Input the optimal number of Principal Components (PCs) -in this work, 7- and a maximum of 100 iterations, and select cross-validation for the validation method. Click "Run".
- Observe the 95% confidence limit, the dashed ring, on the scores plot between PC1 and PC2.
- Mark the sample spectra with scores that occur outside the limit as potential outliers using the "Select Spectra Tool".
- If the marked spectra have high Hotellings T2 values and high Q residuals exclude them from further analysis.
- Click "Analysis | Decomposition" and select PCA.
- Partial Least Squares Regression (PLS-R)
- Click "Analysis | Regression" and select "PLS-R".
- In software 1, right click Y block and select the vector "Parasitemia | Build Model".
- In software 2, input the PLS-R methods. Select the vector "Parasitemia" as the "Y reference" and the sample dataset as "X Data Set" and select cross-validation as the validation method. Click "Run".
- Analyze the regression model. R2 values over 0.80 and root mean square error of cross-validation (RMSECV) values that are less than 0.1% parasitemia are acceptable models.
- Analyze the regression vector and identify the biological bands.
- Click "Analysis | Regression" and select "PLS-R".
- Principal Component Analysis (PCA)
Representative Results
Partial Least Squares (PLS-R) plot and its associated regression vector, Figure 1a and 1b respectively, show that the signal from parasites are distinct enough from the RBCs that they can be used to form a linear regression model to be used for the prediction of parasites in future data sets.
A robust, linear PLS-R model was generated with an R-squared value of 0.87 and a root mean squared cross validation error (RMSECV) of 0.13% parasitemia, Figure 1a. Increasing parasitemia levels are primarily associated with nucleic acid bands, especially 1042, 1083 and 1226/cm, which are assigned to the ν(C=O) from the RNA ribose group, νs(PO4-) and νa(PO4-) respectively, and form the major negative portion of the regression vector. This is because RBCs lack a nucleus and thus nucleic acids are strong indicators for the parasites. Furthermore, the 1226/cm band indicates that the structure has a B-DNA form, which highlights the importance of measuring in the aqueous state12. In contrast, parasites consume RBC components, primarily hemoglobin, and thus a higher protein to lipid ratio is expected in uninfected RBCs as indicated by lipid bands at 2913, 2879 and 1397 cm-1 and 1631 and 1555 cm-1 from the amide I and amide II modes12,13.
The distinct bands clearly show that the signals are true biological bands rather than differentiation based on spurious bands of non-biological origin such as water vapor, which present as narrow and intense bands. Specifically, the presence of DNA bands, Table 1, can be directly attributed to the parasite11,12,13. In general, the regression vector can be interpreted in a similar manner to loadings in PCA. Bands should correlate well with bands and shifts of the original spectra.
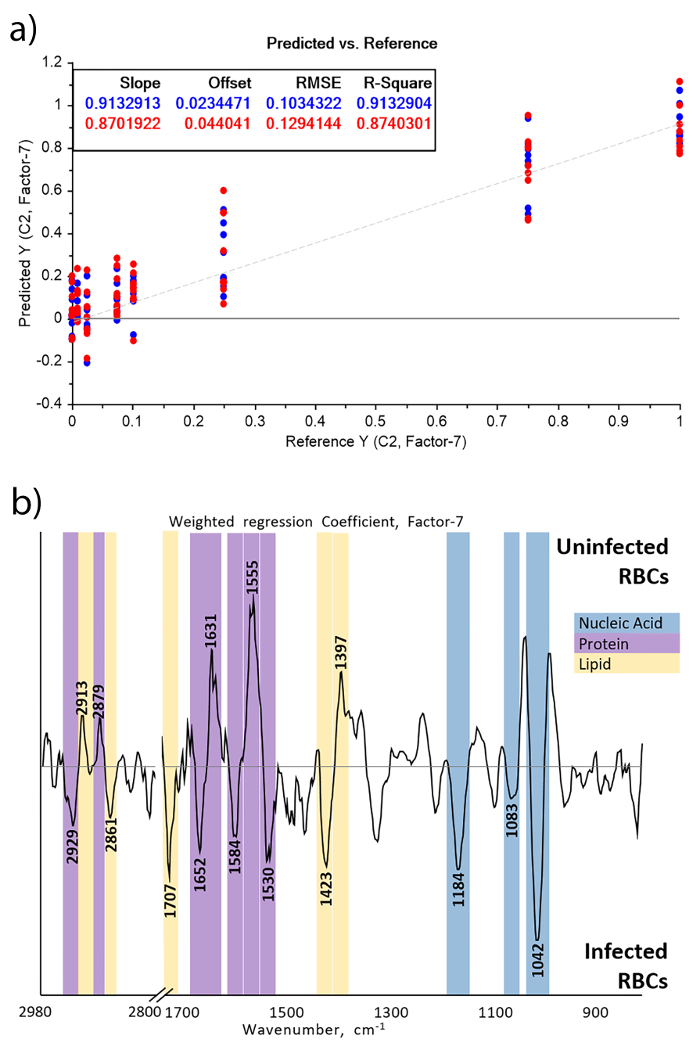
Figure 1: PLS-R regression plot, (a), of RBCs spiked between 0–1% parasitemia and associated regression coefficient, (b). The regression plot demonstrates the ability of the model to predict the parasitemia in each sample, with the known parasitemia on the x-axis and the predicted parasitemia on the y-axis. The regression coefficient describes the contribution of each band to the predictive capability of the model, the more intense the band is the more it contributes to the model and thus how the parasite alters the chemical composition of the blood. Reproduced and modified from Martin, M. et al.10 with permission from the Royal Society of Chemistry. Please click here to view a larger version of this figure.
| Regression Coefficient Band (cm-1) | Assignment25-27 |
| 2929 | νaCH2 from lipids, parasite |
| 2913 | νaCH2 from lipids, red blood cell |
| 2879 | νsCH3 from lipids, red blood cell |
| 2861 | νsCH2 from lipids, parasite |
| 1707 | B-DNA/A-DNA base pair vibration νC=O & νC=N, parasite |
| 1652 | Amide I from α-helix proteins, parasite |
| 1631 | Amide I from β-pleated sheet proteins, red blood cell |
| 1584 | νC=N from imidazoles in Nucleic acids, parasite |
| 1555 | Amide II from proteins, red blood cell |
| 1530 | Amide II, parasite |
| 1423 | B-DNA Deoxyribose, parasite |
| 1397 | ν(COO-) from lipids, red blood cell |
| 1226 | νaPO4- from B-DNA, parasite |
| 1184 | B-DNA Deoxyribose, parasite |
| 1083 | νaPO4- from DNA, parasite |
| 1042 | νC=O from RNA ribose, parasite |
Table 1: Assignments of regression coefficient bands of PLS-R on spectra of malaria spiked RBCs. In PLSR, the parasitemia value associated with a spectrum is computed by multiplying the preprocessed spectrum by the regression vector. Hence, the regression vector depicts the importance and direction of the IR bands in the regression. Because spectra were preprocessed using the second derivative, negative bands are associated with a higher parasitemia while positive bands are associated with a low parasitemia. Reproduced and modified from Martin, M. et al.10 with permission from the Royal Society of Chemistry.
Discussion
PLS-R model is a supervised multivariate method that finds a linear relationship, Y=bX+E, between the predictive variables X (here, the absorbance at each wavenumber) and a continuous variable Y (here, the parasitemia level). In short, the model combines the variables in X to create a new set of latent variables (LVs) that capture the variance on X correlated with Y, and computes a regression vector (b) that multiplied by a new spectrum results in the estimation of the y value. The strength of the model can be taken from two values: the R2 value and the Root Mean Square Error of Cross Validation (RMSECV) value. The former describes the linearity of the model and thus the robustness (the closer it is to 1 the better) and data for models with an R2 value less than 0.8 must be reviewed carefully for outliers and noisy data. The RMSECV value describes the error in which the prediction can be made. It is always best to try and minimize this number as much as possible by making sure to exclude outliers, carefully treating the data and making sure that there are as many biological replicates as possible.
It is thus imperative that good quality data are acquired for building the model. Preparation of the dilution series is critical; specifically making sure that each RBC sample is as homogenous as possible. By using the pipette to draw up and eject the RBCs a few times while swirling or gently flicking the end of the microcentrifuge tube 5–10 times, inconsistencies will be avoided. In a similar vein, it is also critical to clean the crystal between each sample to prevent contamination. Using ultra-pure water and lint free wipes to clean the crystal and checking that the baseline is flat ensures that there are no residues that will contaminate the next sample measurement.
The number of biological samples required for a model that is applicable in the field is at least 30 per condition that is validated with a data set of a similar number. This considerable volume would take a significant amount to generate and thus this is the limitation of the technique. However, as more samples are correctly analyzed they can be added to the predictive model and strengthen it. Thus, once established, the model can become more accurate and can justify the effort needed to set it up.
Depending on the environment and the instrumentation on which measurements are taken, several further issues can arise. If the beamline is not fully enclosed in the instrument, meaning either the joints are not airtight or the ATR window is interchangeable with other sample compartments, ethanol and water vapor can contaminate the spectra. Ethanol appears as a strong sharp band around 1,070/cm 14. This can be avoided by measuring in a space free from ethanol and disinfecting the instrument only once all measurements are taken. Water vapor is caused by changes in the atmosphere and appears as small sharp positive and negative peaks around 1400–1,800 /cm and 3,200–3,600/cm. This can be resolved by increasing the frequency of background measurements and purging the instrument with nitrogen. In cases where purging is unavailable and the water vapor is still quite strong, smoothing algorithms and water vapor compensation can be added to data pre-processing to remove them.
This technique holds a lot of promise for future diagnostics and patient outcomes. Firstly, the method is overall very simple to implement in that the sample needs only be pipetted onto the ATR crystal and then it can be measured directly and be input into the model reducing the potential for user error. This would eliminate the need for highly skilled light microscopist, which currently in some regions of Africa are lacking2. Due to the simplicity, the need for expensive reagents and equipment is also eliminated and thus the diagnostic would be virtually cost-free for patients, which is the major limitation for PCR. This combination would lead to higher numbers of patients coming in for diagnosis much sooner and thus improving patient outcomes. The technique has the potential for extremely high sensitivities that would overcome the low sensitivities of RDTs and would be able to detect asymptomatic carriers sooner and thus can be used as a screening technique. Furthermore, the technique has the potential to be used for other diseases, blood borne or otherwise, where a few microliters or micrograms can be applied to the crystal.
However, in the example model there are many things that need to be addressed before it can be applied in the field. Cross validation is one due to the low number of biological replicates and that is not ideal; rather having a separate validation set to test the model instead is much better. Moreover, the RMSECV is quite high here making the lower levels of quantification unreliable. Increasing the number of replicates will improve this value and must be done before employing this method in the field. This is corroborated in laboratory trials9, where RMSECV was much lower because of the larger sample size. In addition to this, increasing the interval range and points will also improve this value as well. To further improve the diagnostic capability, wild strains of P. falciparum should be used, as the 3D7 P. falciparum is a laboratory strain and may present a different chemical composition due to variations in phenotype.
Declarações
The authors have nothing to disclose.
Acknowledgements
Funding to the authors was provided by the Australian Research Council (Future Fellowship FT120100926 to BRW), National Health and Medical Research Council of Australia (Program grant and Senior Research Fellowship to JGB; Early Career Fellowships JSR; Infrastructure for Research Institutes Support Scheme Grant to the Burnet Institute), and the Victorian State Government Operational Infrastructure Support Grant to the Burnet Institute. We acknowledge Mr. Finlay Shanks for instrumental support.
Materials
| Donor blood | – | – | Must be collected by a trained medical practitioner |
| Stock blood | Australian Red Cross` | – | |
| 3D7 Plasmodium falciparum | The Burnet Institute | – | Laboratory strain wild type equivalent |
| RPMI-1640 media with L-hepes without Sodium Bicarbonate | Sigma Aldrich | R6504 | |
| Albumax (Gibco) | Thermofisher | E003000PJ | Aliquot into 50 mL falcon tubes and store in freezer |
| Sodium Bicarbonate | Sigma Aldrich | S5761 | |
| Giemsa Stain | Sigma Aldrich | 48900 | |
| Sorbitol | Sigma Aldrich | S1876 | Must be filtered before use in culture |
| Blood collection tubes (Vacutainers) | Becton, Dickonson and Company | 367671 | |
| Immersion Oil | Thermofisher | M3004 | |
| 50 mL culture dishes | Falcon | 353025 | |
| 25mL pipette tips | Falcon | 357515 | |
| 10mL pipette tips | Falcon | 357530 | |
| Microscope Slides | Sigma Aldrich | S8902 | |
| Centrifuge tubes 50 mL | Sigma Aldrich | T2318 | |
| Automated pipette controller | Integra-biosciences | 155 015 | |
| Sorvall Legend X1R Centrifuge | Thermofisher | 75004260 | |
| Forma™ 310 Direct Heat CO2 Incubators | Thermofisher | 310TS | |
| Nikon Eclipse E-100 Binocular Microscope | Nikon Instruments | E100_2CE-MRTK-1 | When purchasing ensure that the 100x lens is an oil immersion lens |
| Bruker Alpha Ft-IR Spectrometer with ATR Quick Snap Attachment | Bruker | 9308-3700 | Be sure to request "Eco-ATR" attachment when purchasing |
| Matlab | Mathworks Inc | Multivariate data analysis software | |
| The Unscrambler X | CAMO | Multivariate data analysis software | |
| PLS-Toolbox | Mathworks, Inc. | GUI for Matlab |
Referências
- Hommel, M., Gilles, H., Collier, L., Balows, A., Sussman, M. Chapter 20. Topley & Wilson’s Microbiology and Microbial Infections. Vol. 5 Parasitology. , 361 (1998).
- World Health Organization. . World Malaria Report 2016. , (2016).
- Rogers, W., Murray, P., Baron, E., Pflaller, M., Tenover, F., Yolken, R. Chapter 105. Manual of Clinical Microbiology. , 1355 (1999).
- White, N. J. Chapter 9. Manson’s Tropical Infectious Diseases. 23, 532-600 (2014).
- Lindblade, K. A., Steinhardt, L., Samuels, A., Kachur, S. P., Slutsker, L. The silent threat: asymptomatic parasitemia and malaria transmission. Expert Rev Anti Infect Ther. 11 (6), 623-639 (2013).
- Hanscheid, T., Grobusch, M. How useful is PCR in the diagnosis of malaria. Trends Parasitol. 18 (9), 395-398 (2002).
- Joanny, F., Löhr, S. J., Engleitner, T., Lell, B., Mordmüller, B. Limit of blank and limit of detection of Plasmodium falciparum thick blood smear microscopy in a routine setting in Central Africa. Malar J. 13 (1), 234-241 (2014).
- World Health Organisation. . Malaria Rapid Diagnostic Test Performance: results of WHO product testing of malaria RDTs: round 6 (2014-2015). , (2014).
- Khoshmanesh, A., et al. Detection and quantification of early-stage malaria parasites in laboratory infected erythrocytes by attenuated total reflectance infrared spectroscopy and multivariate analysis. Anal Chem. 86, 4379-4386 (2014).
- Martin, M., et al. The effect of common anticoagulants in detection and quantification of malaria parasitemia in human red blood cells by ATR-FTIR spectroscopy. Anal. 142 (8), 1192-1199 (2017).
- Fabian, H., Mäntele, W. . Handbook of Vibrational Spectroscopy. , (2006).
- Whelan, D., et al. Monitoring the reversible B to A-like transition of DNA in eukaryotic cells using Fourier transform infrared spectroscoptMonitoring the reversible B to A-like transition of DNA in eukaryotic cells using Fourier transform infrared spectroscopy. Nucleic Acid Res. 39 (13), 5439-5448 (2011).
- Wood, B. The importance of hydration and DNA conformation in interpreting infrared spectra of cells and tissues. Chem Soc Rev. 45, 1980-1998 (1980).
- Plyler, E. K. Infrared spectra of methanol, ethanol, and rn-propanol. J Res Natl Bur Stand. 48 (4), (1952).

