Using Human Induced Pluripotent Stem Cell-derived Hepatocyte-like Cells for Drug Discovery
Summary
The protocol presented here describes a platform for identifying small molecules for the treatment of liver disease. A step-by-step description is presented detailing how to differentiate iPSCs into cells with hepatocyte characteristics in 96-well plates, and to use the cells to screen for small molecules with potential therapeutic activity.
Abstract
The ability to differentiate human induced pluripotent stem cells (iPSCs) into hepatocyte-like cells (HLCs) provides new opportunities to study inborn errors in hepatic metabolism. However, to provide a platform that supports the identification of small molecules that can potentially be used to treat liver disease, the procedure requires a culture format that is compatible with screening thousands of compounds. Here, we describe a protocol using completely defined culture conditions, which allow the reproducible differentiation of human iPSCs to hepatocyte-like cells in 96-well tissue culture plates. We also provide an example of using the platform to screen compounds for their ability to lower Apolipoprotein B (APOB) produced from iPSC-derived hepatocytes generated from a familial hypercholesterolemia patient. The availability of a platform that is compatible with drug discovery should allow researchers to identify novel therapeutics for diseases that affect the liver.
Introduction
Success in identifying drugs that can be used to target a rare disease relies on the development of assays that can be used for screening. Hypothesis or target-based screens (reverse pharmacology) are useful, but require a detailed understanding of the molecular basis of the disease. Phenotypic screens (classical pharmacology) avoid the need for a detailed understanding of biochemical pathways, but instead rely on the development of models that accurately mirror the pathophysiology of the disease. Despite enthusiasm for target-based approaches, analyses of FDA approved first-in-class drugs reveal that phenotypic screens have been far more successful1. The overall goal of this method is to establish a platform for high throughput screening that can be used to identify small molecules for the treatment of metabolic liver disease. Several in vitro models have been described including primary hepatocytes, hepatoma cells, and liver progenitor cells2. However, most of these models have limitations, and there is a need for new models that can accurately recapitulate the pathophysiology of metabolic liver deficiencies in culture. Recently, human pluripotent stem cells combined with gene editing have offered an opportunity to model even the rarest of rare diseases in culture without the need to access patients directly3. While the use of patient-specific iPSCs as a tool to discover small molecules for the treatment of rare liver diseases is conceptually reasonable, there are only a few reports demonstrating the feasibility of this approach4. However, we have recently established a platform that used iPSC-derived hepatocytes to successfully identify drugs that can be repurposed for the treatment of deficiencies in liver metabolism5.
This protocol explains the process of differentiating human iPSCs to hepatocyte-like cells in 96-well plates and using them to screen a library of small molecules. It also describes the endpoint analysis using hypercholesterolemia as an example of metabolic liver disease. This approach should be useful to study the role and application of small molecules in the context of infectious liver disease, metabolic liver disease, drug toxicity, and other liver disorders.
Protocol
1. Culture of Human Induced Pluripotent Stem Cells
- Coating recombinant Human E-Cadherin Fc Fusion Protein (E-cad-Fc) or other matrices suitable for hPSC culture6
- Dilute E-cad-Fc to 15 μg/mL with Dulbecco's Phosphate-Buffered Saline containing calcium and magnesium (DPBS (+)).
- Coat 100-mm suspension tissue culture dishes with 5 mL of diluted E-cad-Fc and incubate at 37 ˚C for at least 1 h. Remove substrate and replace with 5 mL of medium (e.g., mTeSR1 referred as M-medium henceforth)7.
Note: The culture medium used in this protocol is prepared following published protocols7. However, several other media preparations have been described, or are available commercially, that are likely compatible with the procedure. - Retrieve a vial of cryopreserved iPSCs from liquid nitrogen. Thaw at 37 ˚C until a small ice crystal remains. Gently pipette cells into a sterile 15-mL conical tube containing 4 mL of the M-medium.
- Centrifuge at 300 x g for 5 min. Remove the supernatant and gently re-suspend cells with 5 mL of the medium supplemented with 10 µM of Y-27632, a selective inhibitor of Rho-associated, coiled-coil containing protein kinase (ROCK).
- Remove the M-medium from step 1.1.2, and transfer 5 mL of cells from step 1.1.4 to E-cad-Fc-coated 100-mm suspension tissue culture dishes.
- Maintaining human iPSCs in culture
- Once iPSCs reach around 80% confluency, remove culture medium and wash once with calcium and magnesium free DPBS (-). Aspirate the DPBS (-) and add a sufficient amount of 0.02% EDTA solution to cover the plates, then incubate for up to 3 min at room temperature.
Note: At 80% confluence, the plate should contain around 2 x 107 cells. It is important not to overgrow the cells, and to ensure that the cells retain an undifferentiated morphology. The pluripotency of the cells can be determined by staining with stem cell marker Tra-1-60 or equivalent8. - As soon as the cells begin to release, remove the 0.02% EDTA solution and flood the dish with 10 mL of the M-medium to release the cells. Aid the detachment of iPSCs by gently pipetting.
Note: The average incubation time for cells to detach is around 3 min, but this needs to be determined empirically for each iPSC line. The cells should be released as small clusters containing around 5 – 10 iPSCs per cluster. - Transfer 1/10 of the suspended cells per fresh E-cad-Fc coated 100 mm suspension tissue culture dish containing 5 mL of the M-medium. Incubate the cultures at 37 ˚C under 4% O2/5% CO2 and change the medium daily.
Note: Although iPSCs are routinely cultured under physiological oxygen conditions to promote pluripotency, iPSCs can also be grown using ambient oxygen.
- Once iPSCs reach around 80% confluency, remove culture medium and wash once with calcium and magnesium free DPBS (-). Aspirate the DPBS (-) and add a sufficient amount of 0.02% EDTA solution to cover the plates, then incubate for up to 3 min at room temperature.
2. Differentiation of Human iPSCs to Hepatocyte-like Cells on 96-Well Plates
Note: This section describes the differentiation of iPSCs in a format that is compatible with screening. Using this approach, it is possible to induce human iPSCs to differentiate and form hepatocyte-like cells in 20 days. While this is suitable for most assays, the length of culture can be extended to improve the maturity of the cells.
- Coating 96-well tissue culture plates with reduced growth factor basement membrane matrix
- Prepare the stock by diluting the matrix to 2 mg/mL with ice-cold sterile DMEM/F12 tissue culture medium. Divide the matrix into 250 μL aliquots and store at -80 ˚C until required. Chill 10 mL of DMEM/F12 on ice for 30 min. Retrieve a 250 μL aliquot of 2 mg/mL matrix stock and thaw on ice.
- Combine the matrix with 10 mL of cold DMEM/F12 by mixing gently using a pre-chilled pipette to make final concentration of 0.05 mg/mL.
- Transfer 100 μL of diluted matrix to each well of a 96-well tissue culture plate. Incubate at 37 ˚C, 4% O2/5% CO2 for at least 1 h. Discard the matrix and replace with 50 μL of M-medium per well. Store at 37 °C, 4% O2/5% CO2 until needed.
- Plate cells for differentiation
- Culture the iPSCs on 100-mm plates until the cells approach 80% confluence. Ensure that the plate contains around 2 x 107 cells. Determine cell numbers by cell flow cytometer or hemocytometer.
Note: With experience, the quality of the iPSCs cells can be judged by microscopy; however, close to 98% of the cells should express pluripotency markers such as the TRA-1-60 epitope when measured by FACS or immunostaining. It is important that the cells remain actively dividing and are not overgrown. - To harvest the iPSCs from each 100-mm plate, aspirate the culture medium and rinse with 5 mL of DPBS (-). Replace the rinse with 3 mL of cell detachment solution (e.g., accutase) and incubate for around 2 min at 37 °C, 4% O2/5% CO2.
Note: It is important to follow the detachment of cells by microscopy. Do not over digest with accutase. The goal is to release the cells as small clusters with minimal treatment. - As soon as the cells begin to detach, harvest by adding 7 mL of the M-medium. Pipette several times to dissociate the cell colonies into small clusters of around 3 – 6 cells (Figure 1A).
- Collect the iPSCs into a 15-mL sterile centrifuge tube and centrifuge at 300 x g for 5 min. Remove the supernatant and re-suspend the cell pellet in 5 mL M-medium.
- Evenly distribute the cells onto 96-well matrix-coated plates by pipetting 50 μL of suspension cells into each well. To set up each 96-well plate, use one 100-mm plate of iPSCs with cells at approximately 80% confluency which contains around 5 x 106 cells to setup the differentiation. Use a multichannel pipette to speed up this process. Culture the cells overnight at 37 ˚C, 4% O2/5% CO2.
Note: It is important that an equal number of cells is deposited into each well to ensure reproducible differentiations.
- Culture the iPSCs on 100-mm plates until the cells approach 80% confluence. Ensure that the plate contains around 2 x 107 cells. Determine cell numbers by cell flow cytometer or hemocytometer.
- Induce differentiation of the cells
- Examine the 96-well plates by microscopy to ensure that cells cover at least 70% of the surface of each individual well. If the coverage is less than 70%, replace the medium with fresh medium and incubate for an additional 24 h at 37 ˚C, 4% O2/5% CO2.
Note: Incubating cells for more than 48 h after plating without inducing differentiation commonly reduces the efficiency of differentiation. Differentiation will also occur using ambient oxygen, although the efficiency may be affected. - For Day 1 to Day 2 of differentiation, replace the culture medium with RPMI supplemented with 2% B27 (without insulin), 100 ng/mL Activin A, 10 ng/mL BMP4, and 20 ng/mL FGF2. Add 100 μL of medium per well and incubate at 37 ˚C, in ambient O2/5% CO2 with a daily medium change for 2 days.
Note: Dealing with large numbers of 96-well plates require the use of an assisted pipette or robot. 12-channel and 96-channel pipettes are ideal for daily medium change. - For Day 3 to Day 5 of differentiation, replace the culture medium with RPMI supplemented with 2% B27 (without insulin) and 100 ng/mL Activin A. Culture cells at 37 ˚C, in ambient O2/5% CO2 with daily medium change for 3 days.
- At differentiation day 5, before replacing with fresh culture medium, treat each well with 100 μL 0.02% EDTA solution for 30 s, then remove and replace with fresh culture medium.
Note: If the differentiation to endoderm is efficient, the monolayer of differentiating cells is prone to peeling from the plate. A brief treatment with 0.02% EDTA solution helps alleviate cell detachment. This treatment does not impact the differentiation to hepatocytes. - For differentiation between Days 6 to 10, replace the culture medium with RPMI/ 2% B27 (with insulin) supplemented with 20 ng/mL BMP4 and 10 ng/mL FGF2 and incubate at 37 ˚C, 4% O2/5% CO2 for 5 days with daily medium change. This stage induces cells to differentiate into hepatic progenitor cells.
- For differentiation between Days 11 to 15, replace the culture medium with RPMI/ 2% B27 (with insulin) supplemented with 20 ng/mL HGF. Incubate at 37 ˚C, 4% O2/5% CO2 for 5 days with daily medium change.
- For differentiation between Days 16 to 20, replace the culture medium with a hepatocyte cell culture medium (HCM) supplemented with 20 ng/mL of Oncostatin-M (OSM). Incubate the cells at 37 ˚C, in ambient O2/5% CO2 with daily medium change for 5 days.
Note: The reproducibility of differentiation between individual wells can efficiently be determined by measuring the relative level of Albumin, AFP, or APOB secreted into the medium by ELISA (see 3.2).
- Examine the 96-well plates by microscopy to ensure that cells cover at least 70% of the surface of each individual well. If the coverage is less than 70%, replace the medium with fresh medium and incubate for an additional 24 h at 37 ˚C, 4% O2/5% CO2.
3. Small Molecule Screening
Note: The following describes the procedure used to identify compounds that can be used to lower the level of APOB in the medium of hepatocyte-like cells derived from familial hypercholesterolemia iPSCs. A description of the model and its use in the identification of cholesterol lowering drugs has been described in detail previously5,9. In the current example, 1,000 compounds from the South Carolina Compound Collection (SC3) library were screened. Each compound was tested at a concentration of 2 μg/mL.
- Treatment of iPSC – derived hepatocytes with small molecules
- Dilute the master compound library to generate plates of working stocks with compounds at a concentration of 20 μg/mL in 2% DMSO and store at -20 ˚C.
- Prepare enough 96-well plates of iPSC-derived hepatocytes to allow each compound to be tested.
Note: The number of compounds to be screened will dictate the number of 96-well plates that are required. In general, a single well should be prepared for each compound, plus additional wells for any control samples. The outermost wells of the plate are commonly avoided to reduce the possibility of edge effects that can corrupt interpretation. Hits can be identified using the z- score method without the need to use replicates. If replicates are included, it is more appropriate to use the t test. - At the completion of differentiation (day 20, see 2.3.7), replace the culture medium with 100 μL of fresh HCM supplemented with 20 ng/mL of Oncostatin-M. Incubate at 37 ˚C, 4% O2/5% CO2 for exactly 24 h. Retrieve the culture medium into a fresh 96-well plate and store at -20 ˚C. Use this sample to calculate the level of APOB in the medium before the addition of drug.
- Add 90 μL of fresh HCM supplemented with 20 ng/mL of Oncostatin-M into each well. Add 10 μL of each compound from the working stock after mixing by pipetting. Add DMSO to 0.2% to control wells to match the final concentration in the treated samples and incubate at 37 ˚C, ambient O2/5% CO2 for exactly 24 h. Collect the culture medium and store at -20 ˚C. Use this sample to determine the level of APOB after treatment with compounds.
- Optional – the remaining cells can be processed for immunostaining, cell viability, or analyses of mRNA levels using standard approaches.
Note: This screen was designed to identify drugs that had a rapid effect on APOB levels. However, depending on the disease model, downstream assays, or mechanism of drug action, it may be beneficial to treat the sample for multiple days.
- Determine APOB levels by ELISA
- Determine Apolipoprotein B (APOB) concentration from both the pre- and post-drug treated samples using a human APOB ELISA development kit following the manufacturer's protocols.
- Preparing the following solutions
- Wash buffer, composed of 0.05% Tween 20 in PBS, pH 7.4. Store at 4 ˚C until use.
- Incubation buffer, composed of 0.05% Tween 20 and 0.1% bovine serum albumin (BSA) in PBS, pH 7.4. Store at 4 ˚C until use.
- Prepare APOB (mAB 20/17) antibody by diluting to 2 µg/mL in PBS, pH 7.4. Add 100 μL of the diluted antibody per well and incubate at 4 ˚C overnight.
- Remove the antibody from the assay plates and wash each well twice with 200 μL of PBS, pH 7.4.
- Add 200 μL/well of 0.05% Tween 20, 0.1% bovine serum albumin (BSA) in PBS, pH 7.4 and incubate on a shaking platform for 1 h at room temperature.
- Prepare an 8-point standard curve using 0.05% Tween 20 and 0.1% BSA in PBS, pH 7.4 with the following concentrations of APOB: 300 ng/mL, 200 ng/mL, 175 ng/mL, 150 ng/mL, 125 ng/mL, 100 ng/mL, 75 ng/mL, and 50 ng/mL.
- Dilute the test samples 1:4 with 0.05% Tween 20 and 0.1% BSA in PBS, pH 7.4.
- After 1 h of blocking with incubation buffer, discard the buffer and wash the assay plates five times with 200 μL of 0.05% Tween 20 in PBS, pH 7.4.
- Completely remove any residual washing buffer and transfer 90 μL of the diluted samples and the APOB standards to the assay plates. Incubate at room temperature on a shaker for 1 h.
- Discard the sample and wash with 200 μL 0.05% Tween 20 in PBS, pH 7.4 per well. Repeat the washes a total of 5 times. Add 100 μL per well of mAB LDL 11-biotin diluted 1:1,000 in 0.05% Tween 20 and 0.1% BSA in PBS, pH 7.4. Incubate at room temperature on a shaking platform for 1 h.
- Discard the reagent and wash with 200 μL per well washing buffer 5 times. Add 100 μL per well of Streptavidin-HRP antibody (1:1,000 dilution using incubation buffer), then incubate at room temperature on a shaking platform for 1 h.
- Discard the reagent and wash with 200 μL 0.05% Tween 20 in PBS, pH 7.4 per well. Repeat the washes a total of 5 times. Completely remove the residual washing buffer from the assay plates and add 100 μL per well of Tetramethylbenzidin (TMB) substrate solution, and incubate at room temperature for 8 min.
- Without removing the TMB substrate, directly add 100 μL per well of stop solution (1N HCl).
- Determine the absorbance at 450 nm using a plate reader.
- A standard curve can be established, using the Four Parameter Logistic Regression method, then used to define the concentration of APOB in each sample10.
- Examine the pre-drug: post-drug ratio of APOB in each sample to identify compounds with the potential to lower APOB levels in the medium.
Representative Results
Generation of hepatocyte–like cells: Figure 1 describes the timeframe of the changes that occur during the differentiation of human iPSCs to hepatocyte-like cells. The culture of iPSCs on E-Cad-Fc provides approximately 2 mm diameter colonies that express the pluripotent marker OCT4 (Figure 1A-B). The morphology of the cells grown on an E-cadherin matrix will be slightly different to those cultured on other surfaces. They tend to have a flattened morphology with a greater cytoplasmic surface area (Figure 1A). Although human pluripotent stem cells can be efficiently cultured on a number of different matrices11, the culture of iPSCs on an E-cadherin matrix is believed to increase the homogeneity of the pluripotent cell population and allows enzyme-free manipulation of the cells during passage12. However, several alternative matrices, including basement membrane matrix, laminins, vitronectin, and mouse embryonic fibroblasts (MEF), can also be used to maintain human iPSCs and all should be compatible with the described procedure13. In all cases, the cell pluripotency can be confirmed by Tra-1-60 or equivalent8.
To initiate the differentiation, the iPSCs should be collected as small clusters (Figure 1A). At the beginning of the differentiation, the plated cells should cover approximately 80% of the surface of the plate. After 5 days of treatment with growth factors, the cells express proteins that are characteristically expressed in the definitive endoderm such as SOX17 and FOXA2 (Figure 1B). At this stage, over 90% of the cells typically express these markers. After 5 more days of differentiation, the endoderm converts to a hepatic fate, and in addition to FOXA2, the cells express HNF4a, which can be identified throughout the remainder of the differentiation process (Figure 1B). As the cells adopt a hepatic fate, they should cover the surface of the plate as a monolayer. After addition of HGF, the cells begin to express proteins that are found in fetal hepatocytes including AFP (Figure 1B). Lipid droplets can be observed within the cytoplasm of the cells. At the completion of differentiation, the cells adopt a cuboidal morphology with a nucleus containing prominent nucleoli and a large cytoplasm with multiple lipid droplets. A subset of the hepatocyte-like cells will be multi-nucleated. Most of the cells express an extensive repertoire of hepatocyte proteins including Albumin (Figure 1B)14.
Effective high throughput screen using hypercholesterolemia as a disease model: Hypercholesterolemia reflects excessive concentrations of Low density lipoprotein-cholesterol (LDL-C) in the serum. In familial hypercholesterolemia (FH), the increased level of serum LDL-C is due primarily to mutations in the Low-density lipoprotein receptor (LDLR) that mediates uptake of LDL-C for clearance by the hepatocytes. We have previously described the generation of iPSCs from a patient with homozygous familial hypercholesterolemia and their use in generating hepatocytes that mirrored the patient's liver disease in culture9. It was demonstrated that these cells could be used successfully as a platform for drug discovery. When a library of ~2,500 drugs was screened, it was found that cardiac glycosides had an unexpected ability to lower LDL-C in iPSC-derived hepatocytes, primary human hepatocytes, and in mice with humanized livers. Moreover, upon examining patient medical records, we found that cholesterol levels fell in patients that were prescribed a cardiac glycoside for the treatment of heart disease. These studies confirmed that iPSC-derived hepatocytes could be successfully used in screens to identify therapeutics.
In order to provide a platform that is compatible with screening, it is important that the differentiations are reproducible and that well to well variation is minimized. Figure 2 shows the results of immunostaining on each well of a 96-well plate to detect HNF4a and Albumin at day 20 of differentiation. As can be seen in the figure, the distribution of these hepatocyte proteins is similar across all wells.
The central protein component of LDL-C is APOB. Here, we have used iPSCs to screen for APOB lowering compounds to provide an example of using iPSC-derived hepatocytes for small molecule screening (Figure 3A). APOB can be easily measured in solution by ELISA. Since ELISA can be performed on large numbers of samples, it can be used as a basis for a screen. The suitability of any assay for high throughput screening is best determined by using a measurement called the Z-factor (or Z')15. This statistical parameter calculates the effectiveness of an assay to distinguish between positive and negative control samples. An assay with a Z-factor >0.5 is considered compatible with a screen15. As can be seen in Figure 3B, the Z' of the APOB assay = 0.73, which confirms the suitability of using the platform for drug discovery.
Identification of compounds that have the capacity to lower APOB levels in the culture medium of iPSC–derived hepatocytes: To determine whether the platform could be used to identify novel compounds that impact APOB levels, we screened 1,000 small molecules from the South Carolina Compound Collection (SC3) Representative Set Lite (RSL). The RSL was generated by the MUSC South Carolina Center for Therapeutic Discovery, and includes compounds that are structurally diverse and reflect the parent library.
The post-drug: pre-drug ratio of APOB was determined for each compound (Figure 3C) and identification of hits was achieved by z-score analysis (Figure 3D)16. In this case, small molecules that reduced APOB with a z-score ≤-2.0 as potential hits were considered. When screening 1,000 small molecules, the number of potential hits with a z-score of ≤-2.0 is usually relatively manageable. However, when conducting a larger screen, we would rely on a more conservative score of ≤-3.0, based on the 3-sigma rule, to increase confidence in potential targets. In this example, we identified 24 potential hits with an APOB ratio that varied from 0.86 to 0.44 (Figure 4A). A secondary assay was performed on quadruplicate samples (n = 4 biological replicates) to exclude compounds that had a negative impact on cell viability and to determine which compounds could reproducibly lower APOB in the medium. Of the original 24 candidate compounds, four were found to be reproducible (p ≤0.05) (Figure 4B-C). Upon further study, 2 of these compounds were found to negatively affect cell viability, suggesting that the observed reduction in APOB was likely to a be a consequence of cell loss. The remaining two compounds would be considered good candidates for follow-up studies.
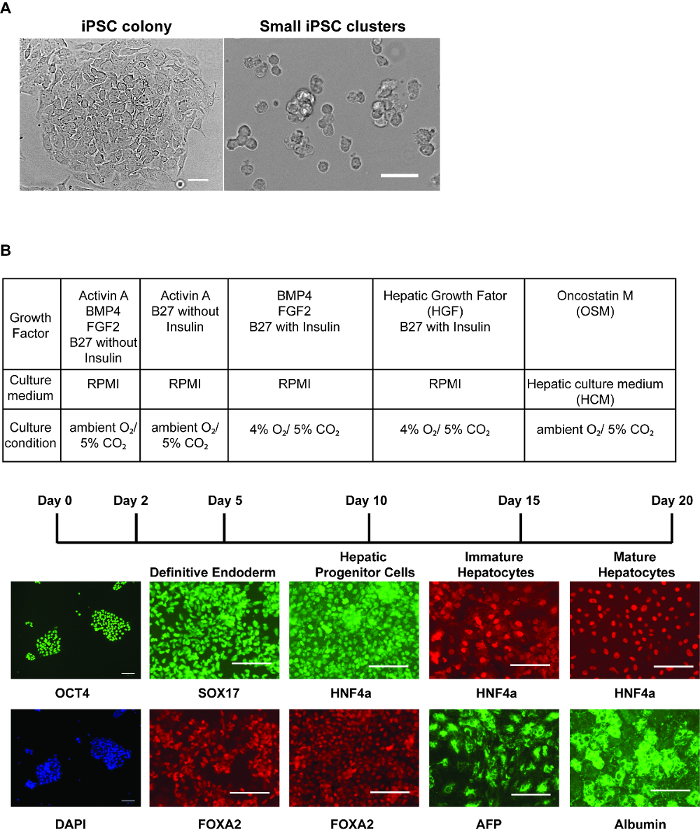
Figure 1: Step-wise hepatic differentiation of human iPSCs. (A) Morphology of iPSC colonies immediately before harvest (left panel) and iPSCs after collecting cells for differentiation (right panel) – after accutase treatment, the cells should be dissociated into 3 – 6 cells/cluster. Scale bar = 50 µm. (B) Table showing culture conditions at each stage of differentiation (upper). Immunostaining (lower panels) was performed to identify marker expression at each stage of differentiation. Panels show OCT4 and DAPI stained nuclei in iPSCs, SOX17 and FOXA2 in definitive endoderm, HNF4a and FOXA2 in hepatic progenitor cells, HNF4a and AFP in immature hepatocyte-like cells, and HNF4a and Albumin in relatively mature hepatocyte-like cells. Scale bars = 100 µm (B). Please click here to view a larger version of this figure.
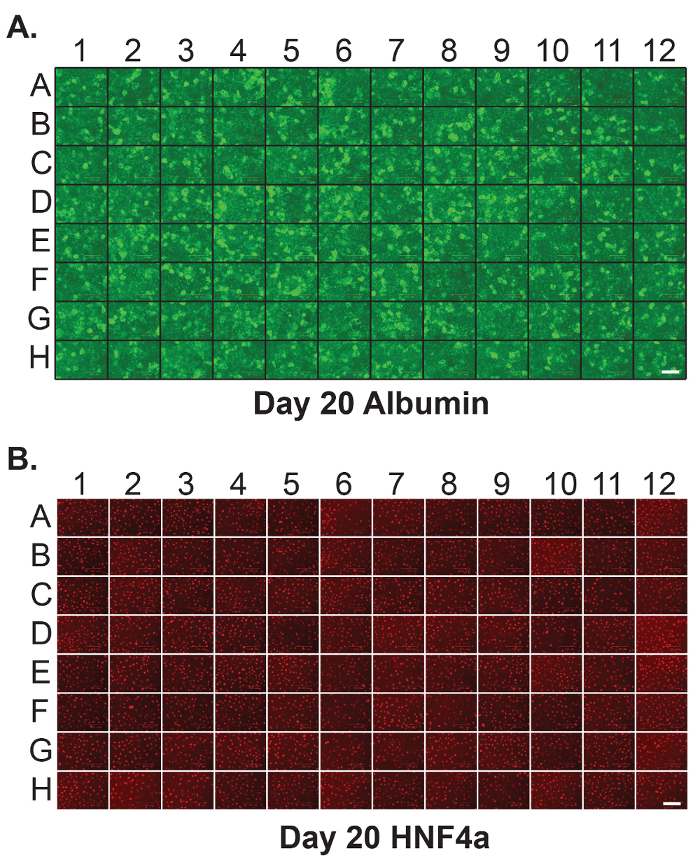
Figure 2: Homogenous hepatic differentiation of human iPSCs in 96-well plates. Immunostaining was performed on each individual well of a 96-well plate containing iPSC-derived hepatocytes at day 20 of differentiation to detect (A) Albumin and (B) HNF4a. Scale bar = 100 µm. Please click here to view a larger version of this figure.
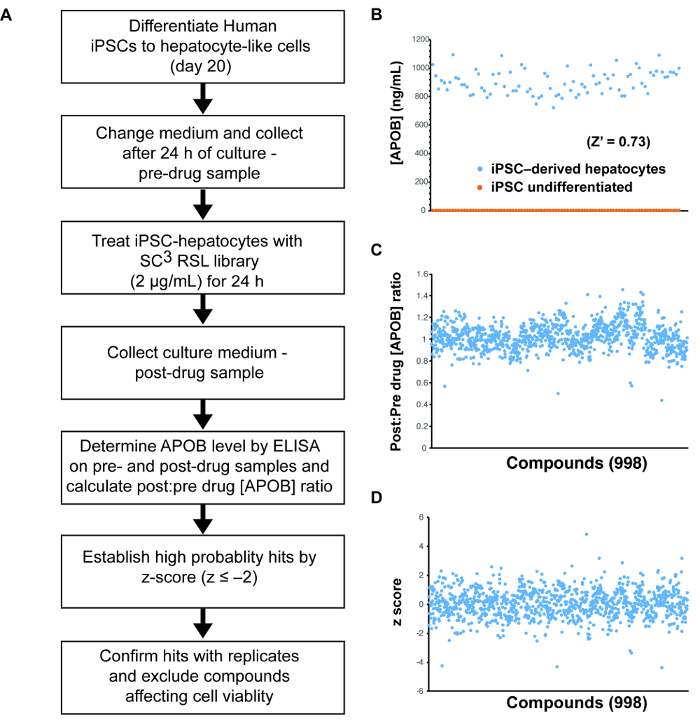
Figure 3: Small molecule screening using human iPSC-derived hepatocytes. (A) Schematic overview of the approach used to identify compounds that can reduce the levels of ABOB in the medium of iPSC-derived hepatocytes. (B) Graph showing theresults of an ELISA assay to detect the level of APOB in the medium of 87 wells (n = 87) of iPSC-derived hepatocytes (blue) and/or undifferentiated iPSCs (orange). These data were used to calculate a Z-factor (Z' = 0.73). (C) Graph showing the post-drug: pre-drug ratios of the concentration of APOB in the culture medium of iPSC-derived hepatocytes treated for 24 h with 998 compounds from the SC3 RSL set. (D) Graph showing z-scores of the data presented in (C). Please click here to view a larger version of this figure.
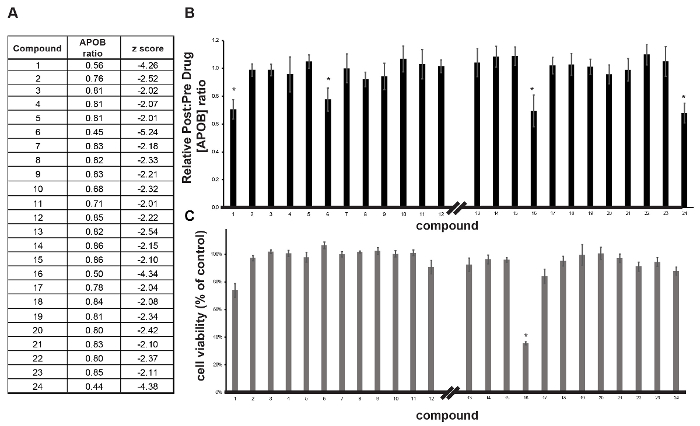
Figure 4: Validation of primary hits. (A) Table showing compounds identified in the primary screen as being able to reduce APOB. (B, C) Bar graphs showing the impact of compounds on post-drug: pre-drug APOB concentration ratio (B) and cell viability (C) in iPSC-derived hepatocytes treated with compounds in follow-up analysis. Error bars reflect ± SEM (n = 4 biological replicates); * p ≤0.05. Please click here to view a larger version of this figure.
Discussion
Target based drug discovery, where small molecules are identified that influence the activity of a specific protein, has been the focus of many existing screening efforts. Although this approach has provided numerous pharmaceuticals, screens based on reversing a phenotype, classical pharmacology, have been more successful in identifying first-in-class compounds that have been clinically efficacious1. A disadvantage to phenotypic drug discovery is that it relies on the availability of appropriate disease models. This can be a challenge when cell models of the disease are unavailable, or when common research animal species fail to recapitulate the clinical symptoms. However, the ability to generate iPSCs from a patient's somatic cells has provided new opportunities to model human disease in culture3. For some diseases, such as rare genetic diseases, patients can be few in number, and difficult to access and consent. However, with the advent of genome engineering, it is relatively straightforward to introduce causative mutations into the iPSC genome, making it possible to model even the rarest of rare diseases.
Once iPSCs are in hand, it is necessary to produce an appropriate cell type in which the symptoms of the disease manifest. For example, diseases affecting neural function may require the generation of neurons or glia17, whereas those affecting the liver may need hepatocytes9,18 or biliary epithelial cells for study19,20,21. This requirement introduces a number of caveats when considering the use of iPSCs for drug discovery. First, it is important to understand the cellular basis of the disease. This can be challenging, especially when cells adopt characteristics based on location or environment. This could be true for hepatocytes that have differences in gene expression profiles and enzymatic function that are dependent upon their location within the liver lobule. Second, working with any cells in isolation can prevent the study of diseases that reflect a disruption to tissue structure such as cirrhosis, cell-cell communication such as neuromuscular disease, or those that are influenced by inflammation such as inflammatory bowel disease.
To study liver disease, several research groups have described protocols to generate hepatocyte-like cells from iPSCs13,22,23,24,25. While most of these protocols are efficient, the limitation of the technique is that cells don't fully recapitulate the function of fresh hepatocytes. This means it is important to determine whether cells generated from iPSCs display the functions necessary to evaluate the impact of a given mutation. Among the genes whose expression does not fully match that found in fresh hepatocytes are those encoding CYP450 proteins25,26. Several of these proteins have roles in drug metabolism and the xenobiotic response. This is important when considering the design of a screen because many drugs require the generation of active metabolites, and this could be affected in iPSC-derived hepatocytes. Efforts to improve the quality of the hepatocytes are being actively pursued27. Despite the caveats, several studies have demonstrated that iPSCs derived from patients with inborn errors in hepatic metabolism can be used successfully to generate hepatocyte-like cells that mirror the pathophysiology of the disease in culture9,18,28,29.
For screens to be successful, it is important to ensure the homogenous differentiation of the hepatocytes from the iPSCs. When differentiations in 96-well plates fail to yield high quality hepatocytes, it is most commonly due either to the poor quality of the parental iPSCs or culture of iPSCs under sub-optimal conditions. Pluripotency of iPSCs must be carefully maintained by culture in a high-quality medium with daily changes as described in protocol 1.2. The density of pluripotent cells in the culture dish should be monitored carefully, because if the cells become over confluent they tend to lose pluripotency and spontaneously differentiate. We also find that culturing the cells in physiological oxygen concentrations promotes pluripotency and is beneficial during some stages of differentiation. However, if appropriate equipment is not available, it is still possible to use ambient oxygen to both culture the pluripotent cells and induce differentiation. We suggest routinely defining the pluripotency of the stock iPSCs by measuring expression of multiple pluripotency markers including OCT-3/4, NANOG, and TRA-1-60 before performing extensive differentiations regardless of culture conditions. We have also observed that when the cells form endoderm at high efficiency, they have a tendency to peal from the surface of the plates as cell sheets. We have found that briefly treating the iPSC-derived endoderm cells with 0.02% EDTA solution can help to reduce cell detachment. Finally, the optimal concentration of cells required for efficient differentiations can differ between iPSC lines and can impact cell detachment. For this reason, it is important to empirically determine the concentration of cells needed for efficient differentiations, especially when working with a new iPSC culture.
In summary, we have described a step-wise differentiation protocol that generates hepatocyte-like cells from iPSCs that are compatible with chemical screens. The hepatocyte-like cells are produced as monolayers in 96-well plates, and the process is efficient and reproducible. We demonstrate the feasibility of screening compounds for their ability to lower the levels of APOB in the culture medium. The differentiation format is compatible with multiple assays including ELISA, qRT-PCR, and high content imaging. We believe that the field will find the approach useful to identify potential therapeutics and for the pharmacological identification of pathways affecting hepatocyte differentiation and function in the future applications5,30.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Institutes of Health (DK55743, DK087377, DK102716 and HG006398 to S.A.D). We would like to thank Dr. Behshad Pournasr, Dr. James Heslop and Ran Jing for their contributions.
Materials
| 100 mm x 20 mm sterile tissue culture dishes | Corning | 430167 | |
| 100 mm x 20 mm sterile suspension culture dishes | Corning | 430591 | |
| 96-wells tissue culture plate | Corning | 3595 | |
| Anti-human Albumin | Dako | A 0001 | |
| Anti-human FOXA2(6C12) | Novus Biological | H00003170-M12 | |
| Anti-human HNF4 alpha | Santa Cruz | SC-6556 | |
| Anti-human Oct-3/4 antibody | Santa Cruz | SC-9081 | |
| Anti-human SOX17 | R&D | AF1924 | |
| Anti-human TRA-1-60 FITC conjugated | Millipore | FCMAB115F | |
| Activin A Recombinant Human Protein | Invitrogen | PHC9563 | |
| B-27 Supplement, minus insulin | Invitrogen | 0050129SA | |
| B-27 Supplement, serum free | Invitrogen | 17504044 | |
| BMP4 Recombinant Human Protein | Invitrogen | PHC9533 | |
| Cell Dissociation Reagent StemPro Accutase | Invitrogen | A1110501 | |
| CellTiter-Glo Luminescent Cell Viability Assay | Promega | 7572 | |
| DPBS+(calcium, magnesium) | Invitrogen | 14040-133 | |
| DPBS-(no calcium, no magnesium) | Invitrogen | 14190-144 | |
| DMEM/F-12, HEPES | Invitrogen | 11330057 | |
| ELISA human APOB ELISA development kit | Mabtech | 3715-1H-20 | |
| Fibroblast Growth Factor 2 (FGF2) | Invitrogen | PHG0023 | |
| Hepatocyte Culture Medium (HCM Bullet Kit) | Lonza | CC-3198 | |
| Hepatocyte Growth Factor (HGF) | Invitrogen | PHC0321 | |
| L-Glutamine | Invitrogen | 25030081 | |
| MEM Non-Essential Amino Acids Solution | Invitrogen | 11140076 | |
| Oncostatin M (OSM) Recombinant Human Protein | Invitrogen | PHC5015 | |
| Penicillin-Streptomycin | Invitrogen | 15140163 | |
| Feeder free pluripotent stem cell medium: mTesR1 | STEMCELL technologies | 5850 | |
| Reduced Growth Factor Basement Membrane Matrix | Invitrogen | A1413301 | |
| RPMI 1640 Medium, HEPES | Invitrogen | 22400105 | |
| StemAdhere Defined Matrix for hPSC (E-cad-Fc) | Primorigen Biosciences | S2071 | |
| TMB-ELISA Substrate Solution | Thermo Scientific | 34022 | |
| Anti-TRA-1-60 FITC conjugated | Millipore | FCMAB115F | |
| Versene (EDTA) 0.02% | Lonza | 17-711E | |
| Y-27632 ROCK inhibitor | STEMCELL Technologies | 72302 |
Referências
- Swinney, D. C., Anthony, J. How were new medicines discovered?. Nat Rev Drug Discov. 10 (7), 507-519 (2011).
- Zeilinger, K., Freyer, N., Damm, G., Seehofer, D., Knospel, F. Cell sources for in vitro human liver cell culture models. Exp Biol Med (Maywood. 241 (15), 1684-1698 (2016).
- Robinton, D. A., Daley, G. Q. The promise of induced pluripotent stem cells in research and therapy. Nature. 481 (7381), 295-305 (2012).
- Lee, G., et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 461 (7262), 402-406 (2009).
- Cayo, M. A., et al. A Drug Screen using Human iPSC-Derived Hepatocyte-like Cells Reveals Cardiac Glycosides as a Potential Treatment for Hypercholesterolemia. Cell Stem Cell. 20 (4), 478-489 (2017).
- Nagaoka, M., et al. E-cadherin-coated plates maintain pluripotent ES cells without colony formation. PLoS One. 1, e15 (2006).
- Ludwig, T. E., et al. Feeder-independent culture of human embryonic stem cells. Nat Methods. 3 (8), 637-646 (2006).
- International Stem Cell Initiative. Characterization of human embryonic stem cell lines by International Stem Cell Initiative. Nat Biotechnol. 25 (7), 803-816 (2007).
- Cayo, M. A., et al. JD induced pluripotent stem cell-derived hepatocytes faithfully recapitulate the pathophysiology of familial hypercholesterolemia. Hepatology. 56 (6), 2163-2171 (2012).
- DeSilva, B., et al. Recommendations for the bioanalytical method validation of ligand-binding assays to support pharmacokinetic assessments of macromolecules. Pharm Res. 20 (11), 1885-1900 (2003).
- Rowland, T. J., et al. Roles of integrins in human induced pluripotent stem cell growth on Matrigel and vitronectin. Stem Cells Dev. 19 (8), 1231-1240 (2010).
- Nagaoka, M., Si-Tayeb, K., Akaike, T., Duncan, S. A. Culture of human pluripotent stem cells using completely defined conditions on a recombinant E-cadherin substratum. BMC Dev Biol. 10, 60 (2010).
- Hay, D. C., et al. Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells. 26 (4), 894-902 (2008).
- Mallanna, S. K., Cayo, M. A., Twaroski, K., Gundry, R. L., Duncan, S. A. Mapping the Cell-Surface N-Glycoproteome of Human Hepatocytes Reveals Markers for Selecting a Homogeneous Population of iPSC-Derived Hepatocytes. Stem Cell Reports. 7 (3), 543-556 (2016).
- Zhang, J. H., Chung, T. D., Oldenburg, K. R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 4 (2), 67-73 (1999).
- Zhang, X. D. Illustration of SSMD, z score, SSMD*, z* score, and t statistic for hit selection in RNAi high-throughput screens. J Biomol Screen. 16 (7), 775-785 (2011).
- Lorenz, C., et al. Human iPSC-Derived Neural Progenitors Are an Effective Drug Discovery Model for Neurological mtDNA Disorders. Cell Stem Cell. 20 (5), 659-674 (2017).
- Rashid, S. T., et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 120 (9), 3127-3136 (2010).
- Lu, W. Y., et al. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat Cell Biol. 17 (8), 971-983 (2015).
- Ogawa, M., et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat Biotechnol. 33 (8), 853-861 (2015).
- Sampaziotis, F., et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat Biotechnol. 33 (8), 845-852 (2015).
- Mallanna, S. K., Duncan, S. A. Differentiation of hepatocytes from pluripotent stem cells. Curr Protoc Stem Cell Biol. 26 (Unit 1G 4), (2013).
- Song, Z., et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 19 (11), 1233-1242 (2009).
- Cai, J., et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 45 (5), 1229-1239 (2007).
- Si-Tayeb, K., et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 51 (1), 297-305 (2010).
- Pashos, E. E., et al. Diverse Population Cohorts of hiPSCs and Derived Hepatocyte-like Cells Reveal Functional Genetic Variation at Blood Lipid-Associated Loci. Cell Stem Cell. 20 (4), 558-570 (2017).
- Davidson, M. D., Ware, B. R., Khetani, S. R. Stem cell-derived liver cells for drug testing and disease modeling. Discov Med. 19 (106), 349-358 (2015).
- Choi, S. M., et al. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology. 57 (6), 2458-2468 (2013).
- Tafaleng, E. N., et al. Induced pluripotent stem cells model personalized variations in liver disease resulting from alpha1-antitrypsin deficiency. Hepatology. 62 (1), 147-157 (2015).
- Jing, R., Duncan, C. B., Duncan, S. A. A small-molecule screen reveals that HSP90beta promotes the conversion of induced pluripotent stem cell-derived endoderm to a hepatic fate and regulates HNF4A turnover. Development. 144 (10), 1764-1774 (2017).

