The Determination of Protease Specificity in Mouse Tissue Extracts by MALDI-TOF Mass Spectrometry: Manipulating PH to Cause Specificity Changes
Summary
Here, we present a protocol to determine protease specificity in crude mouse tissue extracts using MALDI-TOF spectrometry.
Abstract
Proteases have several biological functions, including protein activation/inactivation and food digestion. Identifying protease specificity is important for revealing protease function. The method proposed in this study determines protease specificity by measuring the molecular weight of unique substrates using Matrix-assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) mass spectrometry. The substrates contain iminobiotin, while the cleaved site consists of amino acids, and the spacer consists of polyethylene glycol. The cleaved substrate will generate a unique molecular weight using a cleaved amino acid. One of the merits of this method is that it may be carried out in one pot using crude samples, and it is also suitable for assessing multiple samples. In this article, we describe a simple experimental method optimized with samples extracted from mouse lung tissue, including tissue extraction, placement of digestive substrates into samples, purification of digestive substrates under different pH conditions, and measurement of the substrates' molecular weight using MALDI-TOF mass spectrometry. In summary, this technique allows for the identification of protease specificity in crude samples derived from tissue extracts using MALDI-TOF mass spectrometry, which may easily be scaled up for multiple sample processing.
Introduction
Proteases regulate physiological processes by cleaving proteins, and the initiation of protease activity at specific times and locations is regulated1. It is therefore important to identify the expression and activity of tissues that are locally regulated, and to develop a novel method to detect proteases. The method proposed in this study aims to identify protease specificity in parallel to detecting proteases.
There are some methods for detecting protease specificity. The azocasein method is well-known for its use in the detection of proteases2 but is limited in its ability to obtain further information. Zymography is another comprehensive protease detection method that can be used to determine the molecular weight of proteases3, but it cannot be used to examine substrate specificity. Protease specificity can be determined by spectrometric assays, using substrates such as p-nitroanilide4, and fluorometric assays, using coumarin substrates5 or Fluorescence Resonance Energy Transfer (FRET) assays6. These methods enable single-specificity detection of substrates contained in a single well. In the present study, the protease specificity of tissue extracts is examined under various conditions, as these extracts contain multiple proteases. Protease activity is regulated by several factors, including pH, coenzymes, prosthetic groups, and ion strength. Recently, proteomics-based methods have been used to identify protease specificity; for example, the method reported by Biniossek et al.7 uses the proteome to determine substrate specificity even in crude extracts and allows accurate determination of the cleavage activity of proteases that recognize multiple amino acid sequences8. However, this method is not suitable for the analysis of numerous samples. In contrast, our method enables the simultaneous processing of multiple samples, and the use of Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) mass spectrometry for sample analysis facilitates rapid and easy detection of the cleaved substrates.
This paper outlines a method to evaluate protease specificity by using MALDI-TOF mass spectrometry to measure the molecular weight of unique substrates. The molecular forms of cleaved substrates, along with their theoretical molecular weights, are shown in Figure 1 and Table 1. Previous studies have used substrates containing polyglycine spacers and biotin9; however, these substrates are flawed because the polyglycine sequence may be cleaved by enzymes that recognize the glycine sequence. Additionally, the high affinity between avidin and biotin may lead to a low recovery rate. To improve upon these drawbacks, in this study we synthesized a unique substrate, which was composed of polyethylene glycol (PEG), iminobiotin, and an amino acid that could identify the cleavage site (Figure 1). To discriminate between amino acids of similar molecular weights, D-serine was added between the PEG spacer and the amino acid at the cleaved site.
The N-terminal of the substrate was labeled with iminobiotin, which enables affinity purification from crude samples. The use of iminobiotin instead of biotin is critical; biotin has a strong affinity with avidin, which results in low recovery rates of biotin-labeled substrates from avidin resin, while iminobiotin's affinity for avidin can be altered by pH. Iminobiotin-labeled substrates will bind to avidin in conditions above pH 9, while avidin releases iminobiotin-labeled substrates in conditions below pH 4. Therefore, iminobiotin was used for affinity purification10. In summary, we describe a detailed protocol for detecting protease specificity using unique substrates.
Protocol
Animal experimental protocols were approved by the Nihon Pharmaceutical University's ethics committee and performed in accordance with the guidelines for the Care and Use of Laboratory Animals of the Nihon Pharmaceutical University. The protocol has been adjusted for tissue extracts derived from ICR mice. ICR mice were purchased from Nippon SLC Ltd. (Shizuoka, Japan)
1. Preparation of the Iminobiotin-labeled Substrate
The substrate undergoes solid-phase synthesis using 9-fluorenylmethyloxycarbonyl group (Fmoc)-protected amino acids as described below11. Use Table 2 to determine the Fmoc-compound used, depending on the desired amino acid and stage of synthesis.
- Swill 0.2 g of Fmoc-NH-SAL resin with 10 mL of N,N-dimethylformamide (DMF) in a synthesis column.
- Add 5 mL of 30% piperidine in DMF. Rock the solution for 10 min to cleave the Fmoc-group.
- Check Fmoc cleavage with the trinitrobenzenesulfonic acid (TNBS) test12: Add 10 µL of 1% TNBS in DMF and 10 µL of diisopropylethylamine (DIPEA) to a 10 x 75 mm2 glass tube, then transfer a small amount of resin to the glass tube. The Fmoc-cleaved resin should develop an orange color.
- Repeat steps 1.2-1.3 if this result is negative.
- Wash the resin three times with 5 mL of DMF.
- Add 5 mL of DMF containing 0.3 mmol of the Fmoc-compound listed in Table 2 (starting with the compound used in stage 1), 0.3 mmol of O-(6-Chloro-1H-benzotriazol-1-yl)-N,N,N',N'-tetramethyluronium hexafluorophosphate (HCTU), 0.3 mmol of 1-Hydroxy-1H-benzotriazole, monohydrate (HOBt), and 0.6 mmol of diisopropylethylamine (DIPEA). Rock the solution for 30 min to couple it with the resin.
- Check this step with the TNBS test. If the result is positive, this means the reaction has not sufficiently progressed and this step must be repeated.
- Wash the resin three times with 5 mL of DMF.
- Repeat the elongation reaction by repeating steps 1.2-1.6, changing the Fmoc-compound used in step 1.5 according to Table 2.
- Add 1 mL of 2.5% water, 1% triisopropylsilane (TIS), and 2.5% ethanedithiol (EDT) in trifluoroacetic acid to cleave peptides from the resin and protection groups. Rock the solution for 60 min.
- Filter, and collect filtrate in a 50-mL tube.
- Add 40 mL of cold diethyl ethanol and store overnight at -20 °C.
- Centrifuge 4,000 x g at -20 °C for 30 min. Collect the pellet.
- Use a desiccator for 3 h to remove the diethyl ether.
- Add 1 mL of 50% acetonitrile to water and dissolve the crude peptides. Lyophilize the solution to remove trifluoroacetic acid. Repeat this step several times.
- Purify the peptides using a C18 cartridge column.
- Activate the resin in the C18 cartridge column using 3 mL of acetonitrile (ACN).
- Equilibrate with 5 mL of 5% ACN, 0.1% TFA in H2O.
- Load the crude synthetic peptides into the C18 cartridge column.
- Wash with 10 mL of 5% ACN, 0.1% TFA in H2O.
- Elute the samples with 3 mL of 60% ACN, 0.1% TFA in H2O.
- Check the peptides by measuring their molecular weight using MALDI-TOF mass spectrometry.
- Pipette 1 µL of eluent on a target plate. Immediately add saturated α-cyano-4-hydroxycinnamic acid (CHCA) solution in 50% acetonitrile in 0.1% trifluoroacetic acid
- Crystallize the mixture by air-drying.
- Acquire mass spectra of samples according to the instrument manufacturer's protocol.
- Manually set the MALDI-TOF mass spectrometer to a positive reflect mode.
- Calibrate the mass spectrometer using CHCA, bradykinin fragment 1-7 (monoisotopic molecular weight = 756.3997 Da), and angiotensin II (monoisotopic molecular weight = 1,045.5423 Da).
- Acquire the mass spectra of synthetic peptides, and then evaluate by comparing it with the theoretical molecular weight.
2. Preparation of Tissue Extracts
- Anesthetize male ICR mice in an isoflurane inhalation chamber and verify anesthesia depth via toe pinch. Euthanize the mice by cervical dislocation.
- Using scissors, make a midline abdominal incision through the skin extending up to the xiphoid process.
- Cut through the diaphragm and collect all lung tissue. Cut the tissue into small pieces.
- Wash the tissue three times with ice-cold 50 mM Tris-buffered saline (pH 7.4).
- Weigh the tissues and transfer about 100 mg of tissues to a 12 x 75 mm2 glass tube.
- Add 0.3 mL of the extraction buffer (50 mM Tris-buffer, pH 7.4, containing 0.1% poly (oxyethylene) octylphenyl ether).
- Homogenize the lung tissue samples using a mixing homogenizer.
- Cool the samples on ice.
- Homogenize the samples at 20,000 rpm for 30 s in a blender. Repeat this step until the samples are completely homogeneous.
NOTE: Do not homogenize for more than 1 minute continuously, in order to avoid an increase in temperature.
- Transfer 1,000 µL of the homogenate to a 1.5 mL tube and centrifuge for 5 min at 10,000 x g at 4 °C.
- Transfer 500 µL of the supernatant to a new 1.5 mL plastic tube.
- Use 10 µL of the sample to determine protein concentration, using methods such as the bicinchoninic Acid (BCA) or the Coomassie Brilliant Blue (CBB) method.
- To prevent freezing, add 80% glycerol to the samples for a final concentration of 50% glycerol.
- Store the samples at -80 °C until further use.
3. Digestion of Substrates in Model Protease and Tissue Extracts
- Add 10 µL of 0.1 µg/µL of N-tosyl-L-phenylalanine chloromethyl ketone (TPCK)-trypsin, 0.1 µg/µL of Staphylococcus aureus V8 protease, or 10 µg/µL of tissue extracts into 890 µL of a digestion buffer.
- To clarify the difference in protease specificity depending on the pH, use buffers adjusted to pH 3, 5, 7, and 9. The composition of the digestion buffer is shown in Table 3.
- Carry out a negative control using inactive protease in tissue extracts, prepared by incubation in boiling water (or heating at 100 °C) for 10 min.
- Add 10 µL of iminobiotin-labeled substrates (dissolved in dimethyl sulfoxide (DMSO), final concentration = 100 pmol/10 µL).
- Incubate for 3 hours at 37 °C to digest iminobiotin-labeled substrates.
- After incubation, stop the digestion of substrates with boiling water (or by heating at 100 °C) for 10 min.
- Centrifuge the digested substrates for 5 min at 10,000 x g at 4 °C.
- Transfer the supernatant into a new 1.5 mL tube.
4. Purification of the Iminobiotin-labeled Substrate
- Add 50 µL of 50% streptavidin sepharose beads in 1 M Tris-buffer (pH 9) containing 0.1% poly(oxyethylene) octylphenyl ether.
- Use pH test paper to ensure that the solution is around pH 9. If the pH of the solution is low, adjust it to approximately pH 9 by adding 1 M Tris-buffer (pH 9) containing 0.1% poly(oxyethylene) octylphenyl ether. This step is important, as it involves binding of the iminobiotin-labeled substrate to streptavidin.
- Incubate overnight at 4 °C with continuous gentle mixing on a rotator wheel to bind the iminobiotin-labeled substrates to the streptavidin. The beads should remain suspended throughout the incubation.
- Centrifuge for 1 min at 10,000 x g at 4 °C. Remove the supernatant.
- Wash the beads five times as follows:
- Add 1,000 µL of a wash buffer (50 mM Tris-buffer, pH 9) to the beads.
- Wash for 10 min at 4 °C with continuous gentle mixing on a rotator wheel. The beads should remain suspended throughout the incubation.
- Centrifuge for 1 min at 10,000 x g at 4 °C and remove the supernatant.
- Add 100 µL of 0.2% trifluoroacetic acid to the beads.
- Use pH test paper to ensure that the solution is below pH 2. If the pH of the solution is high, adjust the pH to below 2 by adding 1% trifluoroacetic acid.
- Incubate for 30 min at room temperature with continuous gentle mixing on a rotator wheel.
- Centrifuge for 1 min at 10,000 x g at 4 °C, then transfer the supernatant to a new 1.5 mL tube.
5. Sample Preparation and Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF Mass Spectrometry)
NOTE: All solutions in the following steps should be prepared with high performance liquid chromatography (HPLC)-grade or amino acid sequencing-grade water.
- Pipette 500 µL of acetonitrile onto a C18 resin to activate it. Remove the used acetonitrile. Repeat this step three times.
- Equilibrate the C18 resin with 100 µL of 0.1% trifluoroacetic acid. Remove the eluent. Repeat this step three times.
- To load the samples, bind the samples to the resin using a pipette.
- Wash the resin with 20 µL of 0.1% trifluoroacetic acid. Repeat this step five times.
- Elute the samples with 3 µL of 60% acetonitrile in 0.1% trifluoroacetic acid. Pipette the eluent on a target plate for MALDI-TOF mass spectrometry. Immediately add saturated α-cyano-4-hydroxycinnamic acid (CHCA) solution in 50% acetonitrile in 0.1% trifluoroacetic acid.
- Crystallize the mixture by air-drying.
- Acquire mass spectra of samples according to instrument manufacturer's protocol.
- Manually set the MALDI-TOF mass spectrometer to a positive reflect mode.
- Calibrate the mass spectrometer using CHCA, bradykinin fragment 1-7 (monoisotopic molecular weight = 756.3997 Da), and angiotensin II (monoisotopic molecular weight = 1,045.5423 Da).
- Acquire the mass spectra of samples, paying close attention to the mass spectra from 700 to 900 Da for the cleaved form of the biotin-labeled substrates.
- Identify the detected peaks using Table 1. Detection of the appropriate molecular weight indicates cleavage at the C-terminus of the relevant amino acid.
Representative Results
Evaluation of the method for identification of protease specificity by model protease
The efficiency of this system was evaluated using TPCK-trypsin and V8 protease, whose substrate specificities were obtained. TPCK-trypsin and V8 protease were estimated to cleave the amino acid sequence at the C-terminal of the lysine and arginine residues, and at the carboxy side of the aspartic acid and glutamic acid residues, respectively. Table 1 shows the theoretical molecular weight of the substrate and digested fragments cleaved by specific proteases. Substrate digestion using TPCK-trypsin produced cleaved fragments, two of which could be detected at 839.81 Da and 796.76 Da (Figure 2A). The molecular weights of these fragments were closely in line with the theoretical molecular weights of the fragments cleaved from the C-terminal of lysine and arginine, respectively. In addition, the V8 protease converted precursor substrates to their cleaved form, with molecular weights of 769.78 Da and 755.62 Da (Figure 2B), which matched the theoretical m/z of glutamic acid and aspartic acid, respectively. These results indicate that this method could be used to successfully identify protease specificity with MALDI-TOF mass spectrometry.
Evaluation of substrate specificity at different pH levels using mouse lung extracts
One of the advantages of this technique was that a large number of samples can be simultaneously processed. This study demonstrated that substrate specificity changed according to pH level (Figure 3). In acidic conditions (Figure 3B; pH 5), the molecular weight of a cleaved fragment was 770.13 Da and 826.66 Da. These results indicated that the lung tissue extract contained proteases with the ability to cleave at the C-terminus of glutamic acid and tryptophan. As the pH level gradually increased, the peaks of fragments cleaved by glutamic acid and tryptophan gradually decreased, whereas peaks of fragments cleaved by arginine and lysine gradually increased (Figure 3C, D; pH 7 and 9). These results indicated that the lung extract contained two or more proteases that had different optimum pH and cleavage specificities.
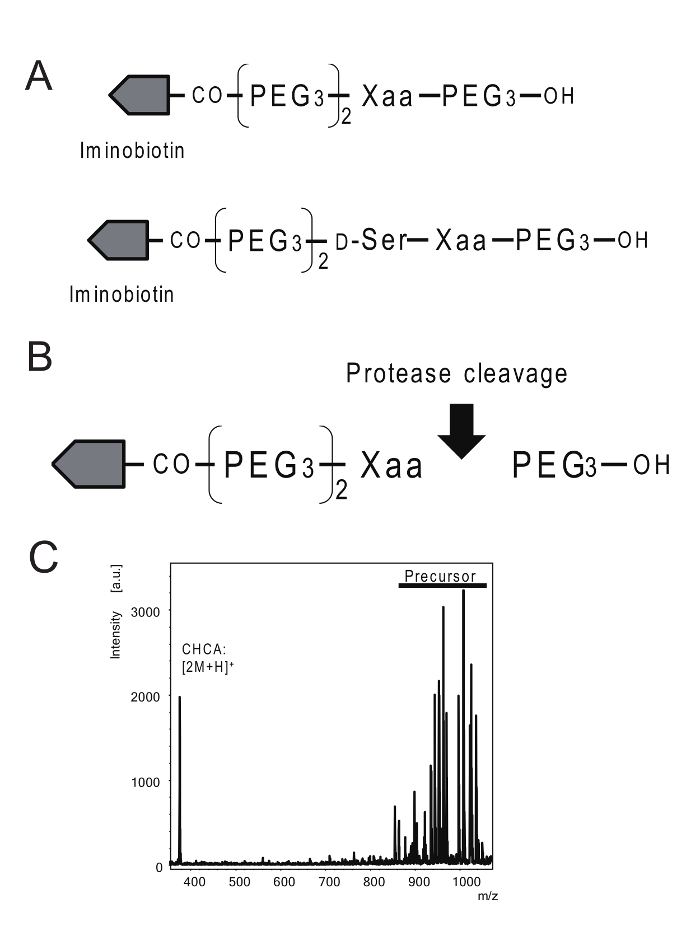
Figure 1: Schema for detection of protease specificity. (A) The structure of iminobiotin-labeled substrate for protease specificity. (B) The substrates had iminobiotin, a polyethylene glycol spacer, and a cleavable amino acid site. (C) The spectra of iminobiotin-labeled substrates. Please click here to view a larger version of this figure.
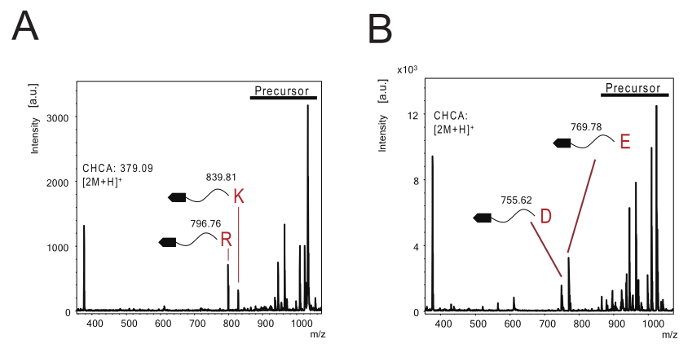
Figure 2: Evaluation of the method to determine protease specificity using a purified enzyme. (A) The spectra for iminobiotin-labeled substrates treated with TPCK-trypsin. The 839.81 and 796.76 peaks matched the theoretical molecular weight of fragments generated by cleavage at the C-terminal side of lysine and arginine, respectively. (B) The spectra for iminobiotin-labeled substrates treated with V8 protease. The 769.78 and 755.62 peaks matched the theoretical molecular weight of fragments generated by cleavage at the C-terminal side of glutamic acid and aspartic acid, respectively. Please click here to view a larger version of this figure.
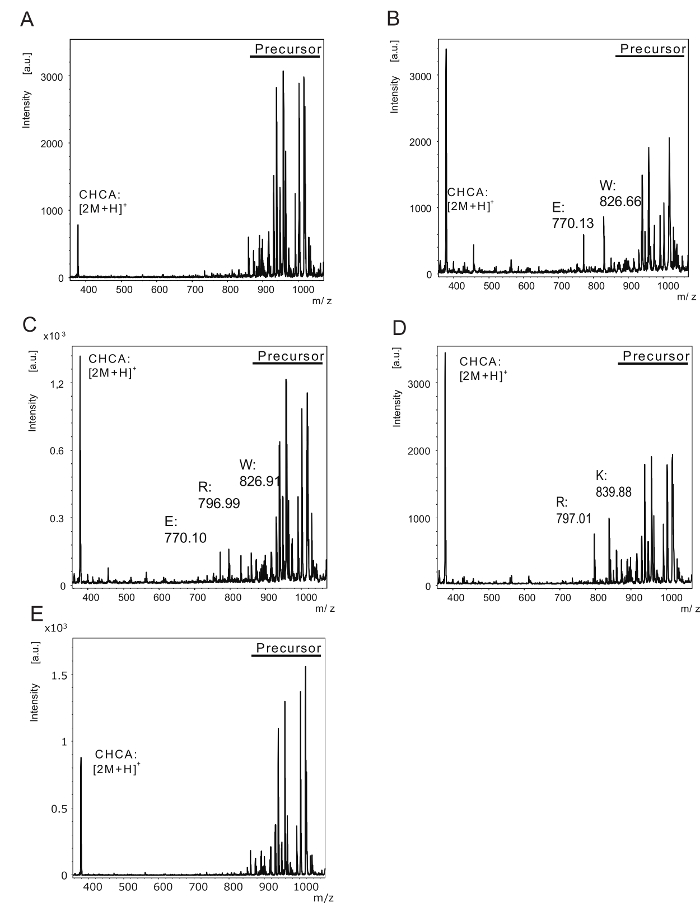
Figure 3: Evaluation of the method to determine protease specificity using tissue extracts. The spectra for iminobiotin-labeled substrates treated with a lung tissue extract. These spectra show peaks obtained at (A) pH 3, (B) pH 5, (C) pH 7, and (D) pH 9. (E) The iminobiotin-labeled substrates incubated with the tissue extract inactivated with heat treatment to evaluate non-specific digestion. Please click here to view a larger version of this figure.
| X | Precursor | Cleaved form |
| G | 886.58 | 697.58 |
| A | 900.60 | 711.60 |
| S | 916.59 | 727.59 |
| P | 926.61 | 737.61 |
| V | 928.63 | 739.63 |
| T | 930.61 | 741.61 |
| C* | 1003.57 | 814.57 |
| I* | 1013.64 | 824.64 |
| L | 942.64 | 753.64 |
| N* | 1014.60 | 825.60 |
| D | 944.59 | 755.59 |
| Q | 957.62 | 768.62 |
| K* | 1028.65 | 839.65 |
| E | 958.60 | 769.60 |
| M* | 1031.60 | 842.60 |
| H | 966.62 | 777.62 |
| F | 976.63 | 787.63 |
| R | 985.66 | 796.66 |
| Y | 992.62 | 803.62 |
| W* | 1015.64 | 826.64 |
| *: D-Ser-Xaa form | ||
Table 1: The theoretical molecular weight of iminobiotin-labeled substrates and the expected molecular weight of protease fragments. Asterisks (*) indicate addition of D-serine between the polyethylene spacer and the cleavable amino acid.
| Xaa: Gly, Ala, Ser, Pro, Val, Thr, Leu, Asp, Gln, Glu, His, Phe, Arg, Tyr | |||
| Stage | Fmoc-compound | ||
| 1 | Fmoc-mini-PEG3-OH | ||
| 2 | Fmoc-Xaa-OH | ||
| 3 | Fmoc-mini-PEG3-OH | ||
| 4 | Fmoc-mini-PEG3-OH | ||
| 5 | Iminobiotin | ||
| Xaa: Cys, Ile, Asn, Lys, Met, Trp | |||
| Stage | Fmoc-compound | ||
| 1 | Fmoc-mini-PEG3-OH | ||
| 2 | Fmoc-Xaa-OH | ||
| 3 | Fmoc-D-Ser(tBu)-OH | ||
| 4 | Fmoc-mini-PEG3-OH | ||
| 5 | Fmoc-mini-PEG3-OH | ||
| 6 | Iminobiotin | ||
Table 2: Sequential list of reagents used for substrate synthesis.
| pH | composition | ||
| 9 | 50 mM Tricine containing 0.1 M NaCl and 10 mM CaCl2 | ||
| 7 | 50 mM Tris-HCl containing 0.1 M NaCl and 10 mM CaCl2 | ||
| 5 | 50 mM sodium acetate containing 0.1 M NaCl and 10 mM CaCl2 | ||
| 3 | 50 mM citric acid containing 0.1 M NaCl and 10 mM CaCl2 | ||
Table 3: Compositions of the buffer solutions used.
Discussion
This protocol uses MALDI-TOF mass spectrometry to identify protease specificity in crude samples derived from tissue extracts and may easily be scaled up for multiple sample processing. In particular, we manipulated pH to cause a change in substrate specificity.
The avidin-biotin complex (ABC) method, which has been widely used in biochemistry due to its binding specificity, was utilized in our protocol. Due to the strong affinity of ABC, the binding of avidin to biotin is almost irreversible. Affinity purification for ABCs is therefore highly inefficient. For this reason, we report that the recovery rate for biotin-labeled substrates is expected to be low. In this study, we only used a low pH level for elution; however, deglycosylated avidin-resin13 or monomeric avidin-resin14 may also be used to purify biotin-labeled substrates.
The method proposed in this study can be adjusted for different experimental systems and applications; bearing in mind that the MALDI-TOF mass spectrometer is unable to conduct quantitative analysis. Once substrate specificity has been determined, it may be better to perform quantification using spectrometric assays, fluorometric assays, or Fluorescence Resonance Energy Transfer (FRET).
The calibration of mass spectrometry was the most important step in this protocol. In cases where substrates with similar molecular weights are detected, the accuracy of the results may be improved by adding the molecular weight of the precursors.
Since the enzyme reaction is enhanced with time, it is possible to extend the reaction time if the enzyme cannot be detected. However, letting the reaction continue for more than 24 h results in side reactions that generate undesired peaks. Therefore, it is advisable to carry out the reaction for up to 18 h or to control testing using heat treated samples. This procedure could determine the protease specificity to within a few femtomol of trypsin.
In conclusion, we introduce a convenient method that uses iminobiotin-labeled substrates to evaluate protease specificity. This method allows for the simultaneous processing of multiple samples and may be used to simplify protease screening.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was supported in part by the Nihon Pharmaceutical University Research Grant from Nihon Pharmaceutical University (2016) and the MEXT KAKENHI Grant Number JP17854179.
Materials
| Fmoc-NH-SAL Resin(-[2,4-Dimethoxyphenyl-N-(9-fluorenylmethoxycarbonyl)aminomethyl]phenoxy resin) | Watanabe Chemical Co., Ltd. | A00102 | |
| N,N-dimethylformamide (DMF) | Watanabe Chemical Co., Ltd. | A00185 | |
| piperidine | Watanabe Chemical Co., Ltd. | A00176 | |
| trinitrobenzenesulfonic acid (TNBS) | WAKO Chemical | 209-1483 | |
| O-(6-Chloro-1H-benzotriazol-1-yl)-N,N,N',N'-tetramethyluronium hexafluorophosphate (HCTU) | Watanabe Chemical Co., Ltd. | A00067 | |
| 1-Hydroxy-1H-benzotriazole,monohydrate (HOBt) | Watanabe Chemical Co., Ltd. | A00014 | |
| diisopropylethylamine (DIPEA) | Watanabe Chemical Co., Ltd. | A00030 | |
| triisopropylsilane (TIS) | Watanabe Chemical Co., Ltd. | A00170 | |
| ethanedithiol (EDT) | Watanabe Chemical Co., Ltd. | A00057 | |
| trifluoroacetic acid (TFA) | Millipore | S6612278 403 | |
| acetonitrile | Kokusan Chemical | 2153025 | |
| C18 cartridge column | Waters | WAT051910 | |
| poly(oxyethylene) octylphenyl ether | WAKO Chemical | 168-11805 | Triton X-100 |
| mixing homogenizer | Kinematica | PT3100 | polytron homogenizer |
| Coomassie Brilliant Blue (CBB) -G250 | Nakarai Chemical | 094-09 | |
| glycerol | Nakarai Chemical | 170-18 | |
| N-tosyl-L-phenylalanine chloromethyl ketone (TPCK)-trypsin | Sigma-Aldrich | T1426 | |
| dimethyl sulfoxide (DMSO) | WAKO Chemical | 043-07216 | |
| streptavidin sepharose | GE Healthcare | 17-511-01 | |
| ZipTipC18 | Millipore | ZTC18S096 | |
| α-cyano-4-hydroxycinnamic acid (CHCA) | Sigma-Aldrich | C2020 | |
| MALDI-TOF mass spectrometry | Bruker Daltonics, Germany | autoflex | |
| 2-iminobiotin | Sigma-Aldrich | I4632 | |
| Fmoc-amino acids | Watanabe Chemical Co., Ltd. |
Referências
- Neurath, H., Walsh, K. A. Role of proteolytic enzymes in biological regulation (a review). Proc Natl Acad Sci U S A. 73 (11), 3825-3832 (1976).
- Surinov, B. P., Manoilov, S. E. Determination of the activity of proteolytic enzymes by means of azocasein. Vopr Med Khim. 11 (5), 55-58 (1965).
- Fernandez-Resa, P., Mira, E., Quesada, A. R. Enhanced detection of casein zymography of matrix metalloproteinases. Anal Biochem. 224 (1), 434-435 (1995).
- Erlanger, B. F., Kokowsky, N., Cohen, W. The preparation and properties of two new chromogenic substrates of trypsin. Arch Biochem Biophys. 95, 271-278 (1961).
- Smith, R. E., Bissell, E. R., Mitchell, A. R., Pearson, K. W. Direct photometric or fluorometric assay of proteinases using substrates containing 7-amino-4-trifluoromethylcoumarin. Thromb Res. 17 (3-4), 393-402 (1980).
- Latt, S. A., Auld, D. S., Vallee, B. L. Fluorescende determination of carboxypeptidase A activity based on electronic energy transfer. Anal Biochem. 50 (1), 56-62 (1972).
- Timmer, J. C., et al. Profiling constitutive proteolytic events in vivo. Biochem J. 407 (1), 41-48 (2007).
- Biniossek, M. L., et al. Identification of protease specificity by combining proteome-derived peptide libraries and quantitative proteomics. Mol Cell Proteomics. 15 (7), 2515-2524 (2016).
- Yamamoto, H., Saito, S., Sawaguchi, Y., Kimura, M. Identification of protease specificity using biotin-labeled substrates. Open Biochem J. 11, 27-35 (2017).
- Hofmann, K., Wood, S. W., Brinton, C. C., Montibeller, J. A., Finn, F. M. Iminobiotin affinity columns and their application to retrieval of streptavidin. Proc Natl Acad Sci U S A. 77 (8), 4666-4668 (1980).
- Merrifield, R. B. Solid Phase Peptide Synthesis. I. The synthesis of a tetrapeptide. J. Am. Chem. Soc. 85, 2149-2154 (1963).
- Hancock, W. S., Battersby, J. E. A new micro-test for the detection of incomplete coupling reactions in solid-phase peptide synthesis using 2,4,6-trinitrobenzenesulphonic acid. Anal. Biochem. 71, 260-264 (1976).
- Marttila, A. T., et al. Recombinant NeutraLite avidin: A non-glycosylated, acidic mutant of chicken avidin that exhibits high affinity for biotin and low non-specific binding properties. FEBS Lett. 467 (1), 31-36 (2000).
- Gravel, R. A., Lam, K. F., Mahuran, D., Kronis, A. Purification of human liver propionyl-CoA carboxylase by carbon tetrachloride extraction and monomeric avidin affinity chromatography. Arch Biochem Biophys. 201 (2), 669-673 (1980).

