A High-Throughput Luciferase Assay to Evaluate Proteolysis of the Single-Turnover Protease PCSK9
Summary
This protocol presents a method to evaluate the proteolytic activity of an intrinsically low-activity, single turnover protease in a cellular context. Specifically, this method is applied to evaluate the proteolytic activity of PCSK9, a key driver of lipid metabolism whose proteolytic activity is required for its ultimate hypercholesterolemic function.
Abstract
Proprotein convertase subtilisin/kexin type 9 (PCSK9) is a single-turnover protease which regulates serum low-density lipoprotein (LDL) levels and, consequently, cardiovascular disease. Although PCSK9 proteolysis is required for its full hypercholesterolemic effect, the evaluation of its proteolytic function is challenging: PCSK9 is only known to cleave itself, undergoes only a single turnover, and after proteolysis, retains its substrate in its active site as an auto-inhibitor. The methods presented here describe an assay which overcomes these challenges. The assay focuses on intermolecular proteolysis in a cell-based context and links successful cleavage to the secreted luciferase activity, which can be easily read out in the conditioned medium. Via sequential steps of mutagenesis, transient transfection, and a luciferase readout, the assay can probe PCSK9 proteolysis under conditions of either genetic or molecular perturbation in a high-throughput manner. This system is well suited for both the biochemical evaluation of clinically discovered missense single-nucleotide polymorphisms (SNPs), as well as for the screening of small-molecule inhibitors of PCSK9 proteolysis.
Introduction
PCSK9 targets the LDL receptor (LDL-R) for degradation, raising LDL cholesterol (LDL-C) and driving atherosclerotic heart disease1,2. Therapeutics targeting PCSK9 robustly lower LDL-C and improve cardiovascular outcomes for patients, even when added to an aggressive lipid-lowering therapy with statins3,4. Currently approved therapies are limited to antibody-based approaches, however, and suffer from a lack of cost-effectiveness5,6. To solve this problem, less costly therapeutic alternatives, a means to identify patients likely to gain greater benefits, or both, are needed.
Small-molecule approaches could target intracellular PCSK9, provide an improved route of administration, and reduce costs, making them the "Holy Grail" in this area7. However, PCSK9 has proven difficult to drug by small molecules. As a protease, targeting PCSK9's proteolytic function is an attractive strategy, as self-proteolysis is the rate-limiting step of PCSK9 maturation8 and is required for its maximal effect on the LDL-R9. To date, however, this strategy has not been successful, likely due to PCSK9's unique biochemistry: PCSK9 cleaves only itself10, performing a single-turnover reaction, and after self-cleavage, the PCSK9 prodomain remains bound in the active site as an auto-inhibitor11, preventing the readout of any further protease activity.
This article presents a method to evaluate PCSK9 proteolytic function in high-throughput fashion8. Through site-directed mutagenesis, investigators can use this assay to probe the effects of coding SNPs found in the clinic to assess them for effects on proteolysis, the rate-limiting step of PCSK9 maturation. Additionally, this method will be useful in the design of high-throughput screens to identify modulators of PCSK9 proteolysis, which are anticipated to ultimately disrupt the presentation of PCSK9 to the LDL-R (and modulate PCSK9's hypercholesterolemic effect). Lastly, this protocol can be adapted to other proteases with intrinsically low activity, provided that i) a specific substrate-protease pair can be found, and ii) a suitable intracellular anchor can be established for the substrate.
Protocol
1. Site-directed Mutagenesis of Protease Vector
- Design and order custom-synthesized oligonucleotides to install a mutation of interest using a modification of standard site-directed mutagenesis protocols12. Standard desalted primers (without additional purification) are perfectly acceptable.
Note: A general approach to primer designs involves creating partially overlapping primers as indicated in Table 1, using a melting temperature (Tm) calculator specific to the polymerase of interest. - Set up polymerase chain reactions (PCRs) for the site-directed mutagenesis on ice, as indicated in Table 2.
- Cycle the PCRs as indicated in Table 3.
- Run 2 – 5 µL of each PCR on a 1% agarose gel and visualize the products to confirm a successful amplification.
Note: A successful PCR will show a single, prominent DNA band at the approximate size marker of the template (~7.8 kB). - Add 0.5 µL (per 25-µL reaction) of DpnI and incubate the samples at 37 °C for 1 h.
- Transform 2 µL of the reaction into chemically competent Escherichia coli.
Note: A fast-growing E. coli strain will reduce incubation times, but any cloning strain will be acceptable. - Plate the transformations onto Luria-Bertani (LB) agar containing 50 – 100 µg/mL carbenicillin. Incubate the plates overnight at 37 °C.
- Select 2 – 4 colonies to grow in a small-scale culture (2 – 5 mL of LB medium with 50 – 100 µg/mL carbenicillin). Incubate the culture at 37 °C at 220 rpm until it is turbid.
- Isolate the plasmid DNA from the cells using a plasmid DNA purification (i.e., “miniprep”) kit according to the manufacturer’s instructions. Quantify the concentration of eluted DNA using a microvolume spectrophotometer.
- Sequence the plasmid DNA extensively using a Sanger sequencing service (such as a core or commercial facility). Prepare the DNA samples according to the service’s specifications. Primers used successfully in prior sequencing reactions are noted in Table 4.
Note: Due to the PCR amplification of the entire plasmid, the sequencing of the entire PCSK9 coding region is recommended for each mutant.
2. High-throughput Luciferase-based Proteolysis Assay
- Cell plating (day 0)
- Use low-passage HEK293T cells for experiments and a culture in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS). Perform all cell work under sterile conditions in a tissue culture hood until ready to assay the luciferase activity. Estimate the number of wells and plates needed for the experiment, anticipating that transfections will be performed in triplicate for each condition tested.
- Dissociate HEK293T cells from a parent flask by treating them with a minimal volume of 0.05% trypsin-ethylenediaminetetraacetic acid (EDTA) to cover the cells. Inactivate the trypsin-EDTA with 2 volumes of DMEM supplemented with 10% FBS and transfer the cells to a sterile tube. Count the cells using an automated cell counter (and staining with trypan blue). Centrifuge the tube at 500 x g for 5 min to recover cells.
- Aspirate trypsin-containing medium and reconstitute the cells in the culture medium to a concentration of 2 x 105 cells/mL. Using a multichannel pipette, transfer 100 µL of the cells to each well of a white (opaque-bottom) 96-well plate, which gives a final seeding concentration of 2 x 104 cells/well.
Note: For initial experiments, it may be useful to additionally seed a sister, clear-bottom 96-well plate, so as to monitor cell growth and adherence during the protocol and guide future troubleshooting. - Incubate the cells at 37 °C and 5% CO2 for 24 h.
- Prepare a master plate of plasmids in a 96-well format. Dilute each plasmid in elution buffer (Tris-HCl, pH 8.5) to 50 ng/µL in an individual well of a 96-well plate.
- Prepare 4 wells each for the positive control (WT) plasmid8, negative control (S386A) plasmid8, and plasmid-free buffer (for mock transfections). If drug or other cellular-based treatments are being planned, then prepare the master plate with the positive control (WT) plasmid in each well, along with 4 wells each of the negative control (S386A) plasmid and the plasmid-free buffer.
- Transfection (day 1)
- Prepare the transfection mixture in a 96-well plate format using deep-well 96-well plates. Perform the transfections in triplicate, being sure to prepare enough reagents to account for pipetting and transfer losses.
Note: The following calculations prepare enough reagent to run one 96-well plate in triplicate (estimated conservatively for 420 wells).- Make a master mix of a diluted lipid transfection reagent, adding 50.4 µL of reagent to 2050 µL of reduced-serum medium. Make a master mix of a DNA precomplexation reagent, adding 33.6 µL of reagent into 1730 µL of reduced-serum medium.
- Using a multichannel pipette, aliquot 16.8 µL of the diluted DNA precomplexation reagent mixture into each well of the deep-well plate.
- Using a multichannel pipette, aliquot 3.2 µL (160 ng) of each plasmid from the master plate into each well of the deep-well plate.
- Using a multichannel pipette, aliquot 20 µL of the diluted lipid transfection reagent mixture to each well of the plate and mix the contents of each well using the multichannel pipette. Cover the plates and let them sit at room temperature (RT) for 10 – 15 min to form lipid:DNA complexes.
Note: Upon transfection, the final components per well will be as follows: 40 ng of DNA, 0.12 µL of transfection reagent, and 0.08 µL of DNA pre-complexation reagent, and the content of each well will be 10 µL in total volume (reduced-serum medium).
- Gently exchange the medium on the 293T cells in the 96-well plates using a multichannel pipette, taking care not to disrupt the cells. Replace the aspirated medium with 95 µL of DMEM supplemented with 10% FBS.
Note: This would be an appropriate time to treat the cells with any drug of interest. - Add 10 µL of the transfection mixture to each appropriate well via a multichannel pipette. Gently swirl the plate to mix the contents in the wells. Incubate the plate at 37 °C with 5% CO2 for 24 h.
Note: The final volume of the wells comes to 105 µL, to account for evaporation over 24 h.
- Prepare the transfection mixture in a 96-well plate format using deep-well 96-well plates. Perform the transfections in triplicate, being sure to prepare enough reagents to account for pipetting and transfer losses.
- Assay (day 2)
- Prepare a stock solution for coelenterazine reagents: 3 M sodium ascorbate [dissolved in phosphate-buffered saline (PBS), prepared fresh], 5 M NaCl, 10 mg/mL bovine serum albumin (BSA; dissolved in PBS, prepared fresh), and 2 mM coelenterazine (dissolved in acidified methanol containing 200 µL of 3 N HCl per 10 mL).
Note: The 2 mM coelenterazine can be stored for 2 weeks when it is kept at -80 °C and in the absence of light. - Prepare 2x coelenterazine reagents for the luciferase readout, with a separate reagent each for the cells and medium, according to Table 5. Mix all reagents save the coelenterazine first, filter the mixture through a 0.22-µm syringe filter, and then add the coelenterazine. Protect the reagents from light until they are ready to be added to the plates.
Note: Due to the loss of solution from filtration, make enough reagent to account for both the loss from filtration, as well as from transfers. Table 5 shows the final concentration of the 2x reagents (as well as the amount of stock solution to add to read out one 96-well plate with one reagent). - Remove the cells from the incubator 24 h after the transfection. Using a multichannel pipette, transfer 50 µL of conditioned medium from each well to a fresh, white-bottomed (opaque) 96-well plate.
- Label the plates as to whether they contain medium or cells. If more than one 96-well plate was transfected, label the plates so as to ensure that each medium-containing plate is paired with its parent plate of cells.
- Using a multichannel pipette, add 50 µL of 2x non-lytic coelenterazine reagent to the plate containing only conditioned medium. Gently rock or shake the plate in the absence of light for 5 – 10 min at RT.
- Using a multichannel pipette, add 50 µL of 2x lytic coelenterazine reagent to the plate containing the cells. Gently rock or shake the plate in the absence of light for 5 – 10 min at RT.
- After the incubation, read out the luminescence of the medium-only plate in a plate reader. Then, read out the luminescence of the cell-containing plate in the same plate reader.
- Discard the plates and save the files for data analysis.
- Prepare a stock solution for coelenterazine reagents: 3 M sodium ascorbate [dissolved in phosphate-buffered saline (PBS), prepared fresh], 5 M NaCl, 10 mg/mL bovine serum albumin (BSA; dissolved in PBS, prepared fresh), and 2 mM coelenterazine (dissolved in acidified methanol containing 200 µL of 3 N HCl per 10 mL).
3. Data Analysis
- Perform an initial data analysis using spreadsheet software. Create a spreadsheet containing the results from the cell and the medium plates.
- Manually inspect the data from the cell plates to identify poorly transfected wells. Wells that show < 5% – 10% of the readout of the negative control (S386A) plasmid should be considered as poorly transfected, making the interpretation of those data suspect.
- Calculate the average background luminescence of each plate from the mock-transfected wells. Subtract the background of each plate from the values of that plate.
Note: This value may be negligible depending on the plate reader used. - Process the data by calculating the proportion of luciferase activity in the medium compared to the overall luciferase activity for each well. Because the cell plate contains both conditioned medium in addition to cells, and the medium-only plate contains the same amount of conditioned medium as the cell plate, it is appropriate to use the following equation:
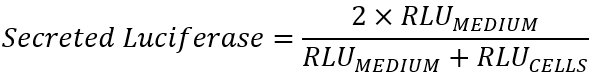
Here, RLU = relative luminescence units, the background-subtracted readout from the plate reader. - Calculate the mean secreted luciferase of the positive control (WT) and the negative control (S386A) wells.
- Evaluate the overall quality of the experiment by calculating a Z-factor13:
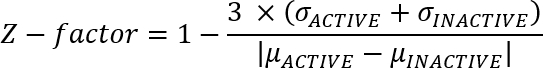
Note: The active values come from the positive control (WT) and the inactive values come from the negative control (S386A) wells. Values closer to 1 indicate a higher experimental quality. Consider repeating the experiment or optimizing the workflow if the value is below 0. - Transfer the data from the spreadsheet program into scientific data analysis software.
- Normalize the secreted luciferase activity to the mean values of the positive control (WT) and the negative control (S386A), setting the positive control as 1 and the negative control as 0.
- Clean the data for outliers using the regression and outlier removal (ROUT) method, setting the maximum false discovery rate to 1%.
- Evaluate for statistically significant differences by comparing the data for each mutant condition (or mutant) to the mean of the WT activity (normalized to a value of 1). Perform multiple unpaired t-tests, correcting for multiple comparisons using the Holm-Sidak method and α = 0.05.
Representative Results
The high-throughput proteolysis assay relies upon overcoming three major challenges. First, to overcome the intrinsically low output of a single-turnover PCSK9 protease, a PCSK9 protease lacking the inhibitory prodomain is used, with the cleavage sequence at the tail of the prodomain linked to a luciferase that can be secreted14. Second, to satisfy the need for the protease to fold in complex with its inhibitory prodomain, the two polypeptides are coexpressed in trans in a cellular context15,16, via a bicistronic vector. Third, to differentiate the cleaved from the uncleaved substrate, the PCSK9 protease is truncated at C678, which prevents the exit of the PCSK9 complex from the endoplasmic reticulum (ER) but has no effect on the proteolytic function17,18. Taken together, this allows for an evaluation of the secreted luciferase as a proxy for the presence and degree of PCSK9 proteolysis (Figure 1A). The general timeframe of the assay is short, with the steps of site-directed mutagenesis of the assay vector (days 1 – 2) followed by the transient transfection (day 3) and luciferase readout (day 4) all completed in as quickly as 4 days, with minimal "hands-on" time (Figure 1B).
Overall, this assay allows for the simultaneous evaluation of PCSK9 proteolysis under a variety of conditions, with the readout done by a luciferase assay. After the acquisition and processing, the data can be visualized in either heat map or tabular format. In particular, mutational analyses of coding SNPs are well suited to this approach. Figure 2 shows a heat map of a saturation mutagenesis library evaluating the cleavage sequence specificity of the PCSK9 protease at substrate sites P6 through P6'. The results show that the optimal cleavage sequence is essentially the same as the WT sequence (SSVFAQ|SIPWNL). In particular, the mutation of the P6 or P4 through P1' residues is poorly tolerated by the protease, consistent with the shallow, hydrophobic binding groove for these positions in the crystal structure of the mature, cleaved PCSK9. From the standpoint of inhibitor development, this narrow sequence specificity profile is a useful finding, as it suggests that no idealized substrate mimetic could outcompete the endogenous cleavage sequence. Figure 3 shows the relative cleavage activity (compared to WT) of a library of missense SNPs described in the clinic, mapped out upon the PCSK9 primary structure. Of the 84 SNPs evaluated, over half showed a significant change in activity compared to the WT protease. These results suggest that alterations in the PCSK9 proteolytic activity are indeed quite common in the clinical population, and such alterations may help to explain the variability in LDL cholesterol levels seen in the clinic.
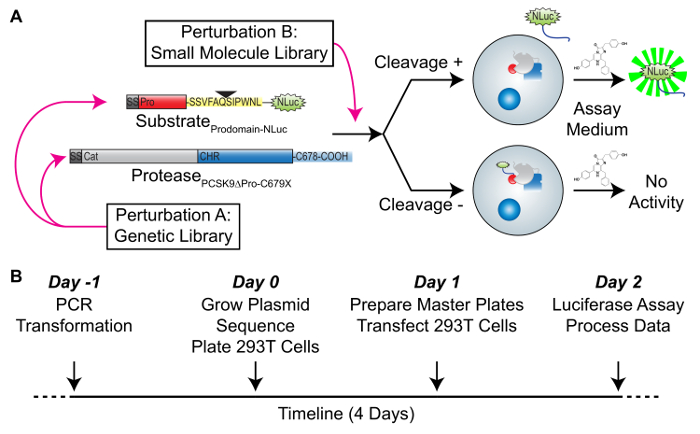
Figure 1: Overall schematic of the high-throughput in trans PCSK9 cleavage assay. (A) This panel shows a biochemical schematic of the assay presented here. The substrate consists of the PCSK9 signal sequence (dark grey) and the prodomain (red) linked to the luciferase that can be secreted (NLuc, green) by the PCSK9 cleavage sequence (yellow). The protease consists of the signal sequence (dark grey), the catalytic domain (light grey), and the cysteine-histidine rich (CHR) domain containing the C679X truncation. The substrate-protease pair is coexpressed at stoichiometric amounts in a 2A-based bicistronic vector, amenable to genetic manipulation. Upon coexpression, the protease and substrate co-fold and remain sequestered in the endoplasmic reticulum (ER). With cleavage activity, the luciferase is freed from the complex and secreted, where it can be detected by the luciferase assay of the conditioned medium. Potential perturbations to the cleavage activity include, but are not limited to, genetic manipulation (perturbation A) or the presence of small molecules (perturbation B). (B) This panel shows the timeline for the overall assay. Site-directed mutagenesis on the WT plasmid is performed on days -1 and 0, followed by the transfection on day 1 and the luciferase assay on day 2, for a total timeline of 4 days. This figure has been adapted from Chorba et al.8. Please click here to view a larger version of this figure.
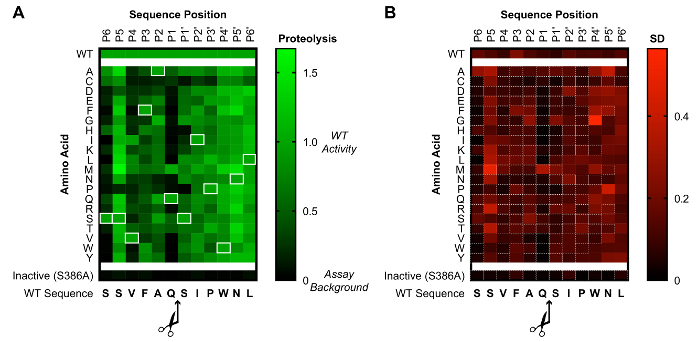
Figure 2: Sequence specificity of the PCSK9 protease. (A) This heat map shows the proteolytic activity, normalized to background and WT, for each single amino acid mutant of a saturation mutagenesis library of the P6 through P6' cleavage sequence. The WT sequence identity is shown at the bottom, with the values for the WT sequence shown both at the top and highlighted by white rectangles. A value of 0 (black) indicates background activity, and a value of +1 (green) indicates the WT value. (B) This heat map shows the standard deviation of the proteolysis values from the same saturation mutagenesis library, with lower values in black and higher values in red. Values outlined in white dashed rectangles represent mutants which showed significantly different proteolysis values from that of the WT protease, as determined by Holm-Sidak-corrected, unpaired t-tests with α < 0.05. The data shown are from 3 independent experiments, each containing 3 replicates. This figure has been adapted from Chorba et al.8. Please click here to view a larger version of this figure.
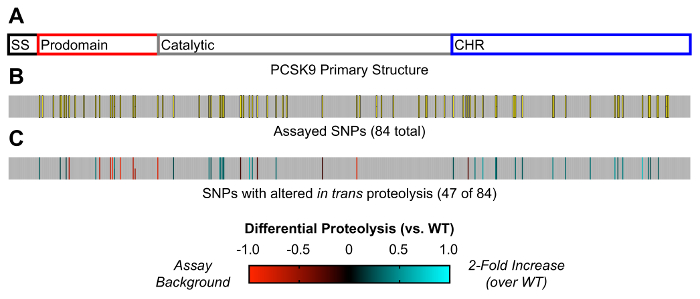
Figure 3: Effects of PCSK9 SNPs on proteolytic function. (A) This panel shows the primary structure of PCSK9, containing the signal sequence (SS; black outline), the prodomain (red outline), the catalytic domain (grey outline), and the cysteine-histidine rich domain (CHR; blue outline). (B) This panel shows the location of 84 clinical SNPs (yellow rectangles with black outlines) placed in the proteolysis assay, mapped onto the primary structure. (C) This panel shows the values of the 47 SNPs with significantly altered proteolytic activity compared to the WT protease, as determined by Holm-Sidak-corrected unpaired t-tests with α < 0.05. The values are mapped onto the PCSK9 primary structure, with a value of -1 (red) indicating no proteolytic activity and a value of +1 (cyan) indicating a 2-fold improvement over the WT protease. This figure has been adapted from Chorba et al.8. Please click here to view a larger version of this figure.
Table 1: Site-directed mutagenic PCR primer design. PCR primers have partially overlapping sequences, according to the design shown. An example of a successful primer pair is shown below the table. Please click here to download this file.
| Component | Stock | Volume (µL) | Final |
| Ultrapure H2O | 17.5 | to 25 µL total volume | |
| Polymerase Reaction Buffer | 5X | 5 | 1X |
| Deoxynucleotides (dNTPs) | 200 µM | 0.5 | 4 µM |
| Template (wild-type (WT)) | 2 ng/µL | 0.5 | 1 ng (total) |
| Primer (Fwd) | 20 µM | 0.625 | 500 nM |
| Primer (Rev) | 20 µM | 0.625 | 500 nM |
| High-Fidelity DNA Polymerase | 2 U/µL | 0.25 | 0.5 U (total) |
Table 2: Components of a PCR for site-directed mutagenesis. The components should be added in the order in which they are listed, with the mixture kept on ice until the reaction begins.
| Step | Temp (°C) | Time | |
| Initial Denaturation | 1 | 98 | 30 s |
| Denaturation | 2 | 98 | 10 s |
| Annealing | 3 | (Tm template)* + 1 | 20 s |
| Extension | 4 | 72 | 30 s/kilobase (kb) (4 min) |
| Cycling | 5 (go to Step 2, 35 cycles total) | ||
| Final Extension | 6 | 72 | 5 min |
| Hold | 7 | 12 | Hold |
| * – Choose the lower Tm template of primer pair | |||
Table 3: Suggested cycling parameters for the PCR. The suggested parameters represent a good starting point but may require optimization.
| Primer | Annealing Location | Sequence (5’ to 3’) |
| 1 | CMV promoter | CGCAAATGGGCGGTAGGCGTG |
| 2 | PCSK9 A95 | TCTCGCAGTCAGAGCGCAC |
| 3 | C-terminus of NLuc | TGTGCGAACGCATTCTGGCG |
| 4 | PCSK9 C301 | GCCAGCGCCTGGCTAGG |
| 5 | PCSK9 E501 | GAGGCCCAAGGGGGCAAG |
Table 4: Validated primers for sequencing. Each primer has been used successfully in sequencing the coding region of the plasmids referenced in this method.
| Component (stock) | Non-Lytic (Medium) | Lytic (Cells) | Amount for 96 wells (1 plate) |
| Sodium ascorbate (3 M) | 300 mM | 300 mM | 700 µL |
| NaCl (5 M) | 5 mM | 5 mM | 7 µL |
| BSA (10 mg/mL, 1%) | 0.10% | – | 700 µL |
| Triton X-100 | – | 0.10% | 7 µL |
| PBS | To 50 µL/well | To 50 µL/well | To 7000 µL |
| Filter through 0.22 µm membrane and transfer 5880 µL of filtrate into fresh tube | |||
| Coelenterazine (2 mM) | 40 µM | 40 µM | 120 µL |
| Total volume: 6000 µL | |||
Table 5: Components of 2x coelenterazine reagents for luciferase readouts. Both non-lytic and lytic reagents are required for each well that is analyzed, so as to separately evaluate the conditioned medium and the transfected cells. The reagents are mixed and filtered prior to the addition of the coelenterazine. After the coelenterazine addition, protect the reagent from light until ready for use.
Discussion
The experimental procedures described above present a method to overcome the intrinsically low activity of the single-turnover protease PCSK9 and evaluate its proteolytic function in a robust manner. The key concept of the assay relies upon converting a single-turnover event into an enzymatically amplified readout. The strengths of the assay include the relatively short time-frame and ease of use of the luciferase reporter, as well as its scalability to high-throughput approaches. In addition, the assay evaluates proteolysis in its native, cellular context. Furthermore, with this assay, clinically identified SNPs can be evaluated for their effects on PCSK9 proteolysis without the need for any human or patient tissue; only knowledge of the genotype of interest is necessary.
Several technical limitations of the assay exist. Though the assay evaluates proteolysis within the cell, it also requires an overexpression of the vector. Because the assay requires the transient transfection of the HEK293T cell line, transfection efficiency adds to the intrinsic well-to-well variability. While the protease-dead S386A PCSK9 serves as the negative control for proteolysis, it also serves as the positive control for the transfection itself, since these cells produce functional, albeit intracellularly trapped, luciferase. Evaluating the raw data of the cellular plates helps to identify those cells with gross variations in transfection efficiency, and these data can be discarded (and the experiment repeated if desired). Furthermore, processing the data for the proportion of secreted luciferase out of the total luciferase produced helps to adjust for smaller variations in both plasmid delivery and transcriptional output. Such variations would be expected to affect the luciferase outputs of the cells and the conditioned medium similarly. Additionally, the vectors used for this assay are amenable to the formation of inducible, isogenic stable cell lines with the commercially available Flp-In T-Rex 293 line, which has been employed with success as a means to bypass the requirement for lipid-mediated transfection. This is likely to be an attractive feature when applying the assay to a small-molecule screening for PCSK9 proteolysis inhibitors. To date, no such inhibitors have been identified, further underscoring the importance of this assay.
The engineering of the PCSK9 protease also creates several biologic limitations. First, the assay measures intermolecular PCSK9 proteolysis, rather than the intramolecular proteolysis that occurs with WT PCSK9. While these two activities generally correlate, it is unlikely that such a correlation exists in all biological situations, and, thus, in some situations, the verification of these effects in an in cis PCSK9 cleavage assay, likely by an alternative method such as immunoblot, may be needed. Additionally, the assay can only evaluate missense SNPs (and not the effect of non-coding variations). Lastly, though PCSK9 proteolysis is the rate-limiting step of PCSK9 secretion, and the reduction of PCSK9 proteolysis is expected to cause a loss-of-function PCSK9 variant (concerning the clinical cholesterol phenotype), this is not true for all SNPs, indicating that at least for some mutations, additional biology is at play8.
The utility of this assay lies in its ability to evaluate the proteolytic function of an intrinsically low-output protease. These design concepts should translate to proteases other than PCSK9 that suffer from similar challenges. To perform this modification, it is necessary to maintain the link between cleavage and luciferase secretion. A strategy where the substrate is replaced with a type I membrane anchor linked to the luciferase via a cleavage sequence specific to the protease of interest should satisfy this requirement.
In summary, this protocol describes a method for evaluating PCSK9 proteolysis in a high-throughput fashion. This method will be useful both for evaluating the effect of clinical SNPs on PCSK9 proteolysis, as well as for the screening of small-molecule inhibitors of PCSK9.
Declarações
The authors have nothing to disclose.
Acknowledgements
The authors thank the generous funding support from the NHLBI/NIH (K08 HL124068 and LRP HMOT1243), NCATS/NIH through the UCSF Clinical and Translational Science Institute Catalyst Program (UL1 TR000004), the UCSF Academic Senate, the Hellman Foundation, a Gilead Sciences Research Scholar Award, a Pfizer ASPIRE Cardiovascular Award (all to John S. Chorba) and the Howard Hughes Medical Institute (to Adri M. Galvan and Kevan M. Shokat).
Materials
| PCR Tubes | USA Scientific | 1402-2900 | For PCR |
| Q5 Hot Start | New England Biolabs | M0493L | High-fidelity DNA Polymerase |
| Deoxynucleotide Solution Mix | New England Biolabs | N0447L | dNTPs (for PCR) |
| pPCSK9-NLucProteaseAssay-WT | Autores | n/a | Available from authors |
| pPCSK9-NLucProteaseAssay-S386A | Autores | n/a | Available from authors |
| Agarose LE | Gold Biotechnology | A-201-100 | For DNA gels |
| E-Gel Imager System with Blue Light Base | ThermoFisher Scientific | 4466612 | For imaging DNA gels |
| SYBR Safe DNA Gel Stain | ThermoFisher Scientific | S33102 | For DNA gels |
| Tris Base | ThermoFisher Scientific | BP152-1 | For DNA gel running buffer |
| Glacial acetic acid | ThermoFisher Scientific | A38-500 | For DNA gel running buffer |
| Ethylenediaminetetraacetic acid solution | Millipore Sigma | 3690 | EDTA, for DNA gel running buffer |
| 1 kb DNA ladder | Gold Biotechnology | D010 | DNA ladder |
| DpnI | New England Biolabs | R0176S | Restriction enzyme |
| LB Agar plates with 100 µg/mL carbenicillin | Teknova | L1010 | LB-Carb plates |
| One Shot Mach1 T Phage-Resistent Chemically Competent E. coli | ThermoFisher Scientific | C862003 | Chemically competent cells |
| LB Broth, Miller | ThermoFisher Scientific | BP1426-2 | LB |
| Carbenicillin | Gold Biotechnology | C-103-5 | Selective antibiotic |
| E.Z.N.A. Plasmid Mini Kit I | Omega BioTek | D6942-02 | DNA Purification Miniprep kit |
| NanoDrop 2000 Spectrophotomer | ThermoFisher Scientific | ND-2000C | Spectrophotometer |
| 293T Cells | American Tissue Culture Collection (ATCC) | CRL-3216 | HEK 293T cells |
| DMEM, high glucose, pyruvate | ThermoFisher Scientific | 11995065 | DMEM, mammalian cell media |
| Fetal Bovine Sera | Axenia Biologix | F001 | FBS |
| Trypsin-EDTA (0.05%), phenol red | ThermoFisher Scientific | 25300062 | Trypsin, for cell dissociation |
| Phosphate buffered saline (PBS) | ThermoFisher Scientific | 10010023 | PBS |
| Countess automated cell counter | ThermoFisher Scientific | C10227 | Automated cell counting |
| Countess cell counting chamber slides | ThermoFisher Scientific | C10228 | Slides for cell counting |
| CELLSTAR Tissue Culture Plates, White, White-Bottom, with Lid | Grenier Bio-One | 655083 | White, white-bottom 96 well plate |
| TempPlate non-skirted 96-well PCR plate, natural | USA Scientific | 1402-9596 | 96 well plate for master plasmid plate |
| Nunc 2.0mL DeepWell Plates | ThermoFisher Scientific | 278743 | 96 well deep well plate |
| Lipofectamine 3000 | ThermoFisher Scientific | L3000008 | Lipid transfection reagent, Lf3K |
| P3000 Reagent | ThermoFisher Scientific | L3000008 | DNA pre-complexation reagent, provided with Lf3K |
| OptiMEM I Reduced Serum Medium | ThermoFisher Scientific | 31985062 | Reduced serum medium for transfection |
| (+)-Sodium L-ascorbate | Millipore Sigma | A4034 | Sodium ascorbate |
| Sodium chloride | Millipore Sigma | S9888 | NaCl |
| Albumin, Bovine Serum, Fraction V, Low Heavy Metals | Millipore Sigma | 12659 | BSA |
| Methanol (HPLC) | ThermoFisher Scientific | A4524 | MeOH |
| Hydrochloric acid | VWR | JT9535-2 | Concentrated HCl |
| Coelenterazine | Gold Biotechnology | CZ2.5 | Luciferase substrate |
| Syringe Filter, Sterile | ThermoFisher Scientific | 09-720-3 | Sterile filter, PVDF, 0.22 µm pore |
| Pipet-Lite Multi Pipette L12-200XLS+ | Rainin | 17013810 | Multichannel pipette |
| Pipet-Lite Multi Pipette L12-20XLS+ | Rainin | 17013808 | Multichannel pipette |
| Pipet-Lite Multi Pipette L12-10XLS+ | Rainin | 17013807 | Multichannel pipette |
| Reagent reservoir | Corning | 4870 | Trough for reagents |
| Centrifuge tubes, 15 mL | ThermoFisher Scientific | 05-539-12 | 15 mL tubes |
| Centrifuge tubes, 50 mL | Corning | 430829 | 50 mL tubes |
| Spark Microplate Reader | Tecan | N/a | Plate Reader |
| Excel | Microsoft | 2016 for Mac | Spreadsheet software |
| Prism | GraphPad Software | v7 | Scientific data analysis software |
Referências
- Park, S. W., Moon, Y. A., Horton, J. D. Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in mouse liver. Journal of Biological Chemistry. 279 (48), 50630-50638 (2004).
- Cohen, J. C., Boerwinkle, E., Mosley, T. H., Hobbs, H. H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. New England Journal of Medicine. 354 (12), 1264-1272 (2006).
- Ridker, P. M., et al. Cardiovascular Efficacy and Safety of Bococizumab in High-Risk Patients. New England Journal of Medicine. 376 (16), 1527-1539 (2017).
- Sabatine, M. S., et al. Evolocumab and Clinical Outcomes in Patients with Cardiovascular Disease. New England Journal of Medicine. 376 (18), 1713-1722 (2017).
- Kazi, D. S., et al. Cost-effectiveness of PCSK9 Inhibitor Therapy in Patients With Heterozygous Familial Hypercholesterolemia or Atherosclerotic Cardiovascular Disease. Journal of the American Medical Association. 316 (7), 743-753 (2016).
- Kazi, D. S., et al. Updated Cost-effectiveness Analysis of PCSK9 Inhibitors Based on the Results of the FOURIER Trial. Journal of the American Medical Association. 318 (8), (2017).
- Pettersen, D., Fjellström, O. Small molecule modulators of PCSK9 – A literature and patent overview. Bioorganic & Medicinal Chemistry Letters. 28 (7), 1155-1160 (2018).
- Chorba, J. S., Galvan, A. M., Shokat, K. M. Stepwise processing analyses of the single-turnover PCSK9 protease reveal its substrate sequence specificity and link clinical genotype to lipid phenotype. Journal of Biological Chemistry. 293 (6), 1875-1886 (2018).
- Maxwell, K. N., Breslow, J. L. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proceedings of the National Academy of Sciences of the United States of America. 101 (18), 7100-7105 (2004).
- Benjannet, S., et al. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. Journal of Biological Chemistry. 279 (47), 48865-48875 (2004).
- Cunningham, D., et al. Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia. Nature Structural & Molecular Biology. 14 (5), 413-419 (2007).
- Liu, H., Naismith, J. H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnology. 8, 91 (2008).
- Zhang, J., Chung, T., Oldenburg, K. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. Journal of Biomolecular Screening. 4 (2), 67-73 (1999).
- Hall, M. P., et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chemical Biology. 7 (11), 1848-1857 (2012).
- McNutt, M. C., Lagace, T. A., Horton, J. D. Catalytic activity is not required for secreted PCSK9 to reduce low density lipoprotein receptors in HepG2 cells. Journal of Biological Chemistry. 282 (29), 20799-20803 (2007).
- Chorba, J. S., Shokat, K. M. The proprotein convertase subtilisin/kexin type 9 (PCSK9) active site and cleavage sequence differentially regulate protein secretion from proteolysis. Journal of Biological Chemistry. 289 (42), 29030-29043 (2014).
- Benjannet, S., Rhainds, D., Hamelin, J., Nassoury, N., Seidah, N. G. The proprotein convertase (PC) PCSK9 is inactivated by furin and/or PC5/6A: functional consequences of natural mutations and post-translational modifications. Journal of Biological Chemistry. 281 (41), 30561-30572 (2006).
- Zhao, Z., et al. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. American Journal of Human Genetics. 79 (3), 514-523 (2006).

