Measurement of BK-polyomavirus Non-Coding Control Region Driven Transcriptional Activity Via Flow Cytometry
Summary
In this manuscript, a protocol is presented to perform FACS-based measurement of BK-polyomavirus transcriptional activity by using HEK293T cells transfected with a bidirectional reporter plasmid expressing tdTomato and eGFP. This method further allows to quantitatively determine the influence of novel compounds on viral transcription.
Abstract
Polyomaviruses, like the BK-polyomavirus (BKPyV), can cause severe pathologies in immunocompromised patients. However, since highly effective antivirals are currently not available, methods measuring the impact of potential antiviral agents are required. Here, a dual fluorescence reporter that allows the analysis of the BKPyV non-coding control-region (NCCR) driven early and late promoter activity was constructed to quantify the impact of potential antiviral drugs on viral gene expression via tdTomato and eGFP expression. In addition, by cloning BKPyV-NCCR amplicons which in this protocol have been exemplarily obtained from the blood-derived DNA of immunocompromised renal transplanted patients, the impact of NCCR-rearrangements on viral gene expression can be determined. Following cloning of the patient derived amplicons, HEK293T cells were transfected with the reporter-plasmids, and treated with potential antiviral agents. Subsequently, cells were subjected to FACS-analysis for measuring mean fluorescence intensities 72 h post transfection. To also test the analysis of drugs that have a potential cell cycle inhibiting effect, only transfected and thus fluorescent cells are used. Since this assay is performed in large T Antigen expressing cells, the impact of early and late expression can be analyzed in a mutually independent manner.
Introduction
Polyomaviruses represent an independent family of small double-stranded DNA (dsDNA) viruses with the Simian virus 40 (SV40) as a prototype species. The primary infection mainly occurs during the childhood, which usually proceed without disease symptoms and usually cause latent infections in immune competent hosts. The BK-polyomavirus (BKPyV) mainly persists in renal tubules cells without causing nephropathologies, however, in case of impairment of the immune-competence after renal transplantation the virus can reactivate and cause severe damages and impaired graft function when reaching a high viremia (1 x 104 BKPyV DNA copies/mL)1,2. In approximately 10% of kidney transplant recipients, reactivation of the BK-polyomavirus (BKPyV) results in a polyomavirus associated nephropathy (PyVAN), which was up to 80% associated with a high risk of renal allograft failures3,4. Since no approved antiviral agents are available, current therapy is based on the reduction of immunosuppression. Interestingly, mTOR inhibitors seem to have an antiviral effect; thus, switching the immunosuppressive therapy to mTOR-based immunosuppression might represent an alternative approach to prevent progression of the BKPyV viremia5,6,7. However, the mTOR-based antiviral mechanism is currently still incompletely understood. Thus, methods measuring the impact of potential antiviral agents in clinically relevant concentrations are required.
The circular genome of the BKPyV consists of approximately 5 kb harboring a non-coding control region (NCCR) that serves as an origin of replication and concomitantly a bidirectional promoter driving the expression of early and late phase mRNA transcripts. Since spontaneously occurring NCCR-rearrangements, deletions, and duplications are found in pathogenic BKPyV8 and significantly accumulated in patients suffering from PVAN5,9, a comparison of archetypical (wt) and re-arranged (rr) NCCR-activities are helpful to characterize viral replicative fitness.
As summarized in Figure 1, this protocol describes a commonly used method to measure BKPyV NCCR transcriptional activity by quantifying the fluorescence of two fluorophores tdTomato and eGFP expressed from a reporter plasmid5,9,10,11. The procedure is performed in the presence of the SV40 large T antigen (lTAg), which allows to analyze the impact of potential antiviral agents on the early and late NCCR-activity separately5. This assay further analyzes the impact of rearrangements on the NCCR activity and comparison with wt-NCCRs5,9. The reporter plasmid harbors the SV40 late polyadenylation signal downstream of each fluorophore open reading frame to ensure comparable and efficient processing of both transcripts for tdTomato and eGFP, respectively. Compared to qRT-PCR based methods5,12, this FACS-based approach represents a low cost and high throughput compatible alternative since no complicated extraction protocols for infected cell culture and no expensive antibodies for immune fluorescence staining are needed. Furthermore, since a defined amount of fluorescent cells are analyzed via flow cytometry, the analysis of cell cycle inhibiting agents is also possible in a quantitative manner.
Protocol
This protocol follows the guidelines of human research as approved by the ethic committee of the medical faculty of the University of Duisburg-Essen (14-6028-BO).
1. Collection of blood or urine samples and isolation of polyomavirus DNA
- Collect at least 3 mL of blood in EDTA tubes or urine in acquisition tubes.
- Centrifuge the sample at 2,500 x g for 15 min. If needed, pipette plasma into a new tube and store the plasma samples at 4 °C for several days or freeze at -20 °C for longer storage.
- Prepare 40 µL of proteinase K into a 1.5 mL microcentrifuge tube and add 400 µL of plasma by pipetting.
- Lyse the sample by adding 400 µL of lysis buffer, vortex for 15 s, and incubate for 10 min at 56 °C.
- Isolate the DNA using a DNA blood extraction kit as described in the manufacturer's instructions. Briefly, add alcohol and load the lysates onto a spin column containing a DNA binding silica-based membrane. Wash the columns several times to yield pure DNA.
- Elute the yielded DNA in 30-50 µL of TE buffer.
2. Amplification of the non-coding control region (NCCR)
- Perform the following procedure in physically separated rooms. Use an isolated room to prepare the reagents and distribute the mix to the PCR tubes.
- Prepare the Master Mix for the pre-PCR using primer pair A (Table 1) in a total volume of 50 µL and use the previously isolated DNA from step 1.6. The pipetting scheme for preparing the master mix for the pre and nested PCR is printed in Table 2.
- Distribute 45 µL of the master mix into the PCR tubes.
- Add 5 µL of the isolated DNA into the PCR tubes.
NOTE: Use another room than the one for the master mix preparation to avoid contamination. - Run the PCR with the reaction conditions as illustrated in Table 3. Repeat denaturation, annealing, and extension in 35 cycles.
- For the nested amplification use primer pair B harboring the restriction sites for AgeI and SpeI (Table 1).
- Run the nested PCR with the reaction conditions as described in steps 2.2 to 2.5 using 5 µL of the pre-PCR.
- Mix 10 µL of the PCR product with 2 µL of 6x gel loading dye. Load 10 µL of the mix on a 1.5% agarose gel, and run the gel for 30 min at 60 mA.
- Visualize the gel using an appropriate UV documentation system.
NOTE: The size of the amplicon is expected to be between 300-500 bp depending on whether rearrangements (deletions, insertions, or duplications) occurred (Figure 2C). - Purify the PCR amplicons using a PCR purification kit according to the manufacturer's instructions. Elute the PCR amplicons in 30 µL of elution buffer.
- Send the amplicons for Sanger sequencing using primer pair B.
3. Cloning of the NCCR into the dual fluorescence reporter
- Digest the purified amplicons (step 2.10) with AgeI and SpeI for 2 h at 37 °C as described in Table 4.
- Repeat step 2.10 and purify the digested amplicons.
- In parallel, also digest the plasmid backbone (1.5 µg) with AgeI and SpeI for 2 h at 37 °C as described in Table 5. This step only needs to be performed once. For subsequent reactions, freeze the digestion at -20 °C.
- Analyze the digested plasmid backbone on a 0.8% low melt agarose gel.
NOTE: Do not run the gel with a current higher than 40 mA. The expected insert of the spacer region originally derived from the plasmid pEX-K4 2-LTR CD313 is 128 bp (Figure 2C). - Visualize DNA fragments using long-wave UV light (320 nm) and cut out the backbone band using a clean scalpel. Transfer the low melt gel fragment into a new 1.5 mL microcentrifuge tube. Avoid long UV exposure times to prevent DNA damage.
NOTE: Since low melt agarose is used, it is not necessary to purify the DNA before ligation. - Heat the backbone containing the low melt agarose piece for 10 min at 65 °C in order to melt the gel piece and mix every 2 min by gentle vortexing. The melted gel can be directly used for ligation.
- Ligate the backbone and the digested amplicon using T4 DNA ligase over night at 16 °C using the scheme shown in Table 6.
- Perform transformation in E. coli using the heat shock method, plate bacteria on LB-amp plates, and culture at 37 °C over night.
- On the next day, select three positive clones and prepare overnight E. coli cultures (5 mL of LB-Amp) and culture at 37 °C.
- Isolate the plasmid-DNA using standard protocols as described elsewhere14, perform AgeI and SpeI digestion to check for positive clones and visualize cut out fragments on a 1% agarose gel. The spacer band (128 bp) will be replaced by the larger NCCR-sequence (300-500 bp, see above).
- Send the plasmid for Sanger sequencing using primer EGFP-N or primer pair B (Table 1).
- Since mutations might spontaneously occur, compare the sequencing results with the sequencing results obtained with the amplicon. Only use the clones containing identical sequences compared to the amplicon.
- Use a molecular workbench software (e.g., freeware GENtle 1.9.4 or other sequence editing programs) to align and edit the obtained sequences (Figure 3).
- Start GENtle 1.9.4 and import the DNA sequences by clicking on the Import button (green down arrow).
- Next click on Tool and select Alignment from the menu (Use Ctrl+G as a shortcut) and choose the DNA sequences for alignment.
- Add a sequence to the alignment by clicking on Add or remove a sequence by clicking on Remove. Include an archetypical NCCR consensus sequence like JN19243815, do not use the NCCR-sequence from the commonly used Dunlop-strain16, since it harbors rearrangements and duplications others than the archetypical BKPyV strains.
NOTE: The NCCR already contains the translational start codon (Figure 3). - Choose a method for the alignment by clicking on Algorithm in the toolbar and choose clustal W. Set the alignment parameters to match 2; gap extension penalty -1; gap penalty -2 and click on OK to run the alignment.
4. Transient transfection of HEK293T cells with the reporter plasmid and treatment with potential antiviral agents
- To prepare sufficient amounts of plasmid DNA for subsequent transfection experiments prepare a 150 mL overnight culture. Isolate the plasmid DNA using a plasmid isolation kit. Alternatively, other plasmid purification kits may be used.
- Seed 1 x 105 HEK293T cells in DMEM containing 10% FCS, penicillin and streptomycin (1x) per well of a 12-well plate 24 h prior to transfection and incubate overnight at 37 °C and 5% CO2 to maintain active proliferation during transfection.
NOTE: Cells should be approximately 80% confluent at transfection. - Place 250 µL of reduced serum media in a sterile tube and add 1 µg of each reporter plasmid DNA and mix gently by pipetting.
- Add 3 µL of the transfection reagent to the DNA mixture, mix gently by pipetting and incubate for 15 min. Pre warm the transfection reagent to the ambient temperature of 22 °C and vortex gently before use.
- Add the mixture drop-wise to the wells and gently distribute to the well.
- After 4 h, replace the supernatant with fresh medium containing the testing agents and solvent control. Incubate at 37 °C until analysis. In this example, the mTOR inhibitors INK128 (100 ng/mL) and rapamycin (100 ng/mL) were used.
5. Fluorescence microscopy and flow cytometry
- Check cells for the red and green fluorescence under the fluorescence microscope (Figure 4).
NOTE: Red and green fluorescence correspond to the early and late BKPyV gene expression, respectively. - After 72 h post transfection, aspirate the supernatant and wash the cells twice with 1 mL of cold PBS.
- Gently add 500 µL of trypsin and turn the plate slightly to avoid premature detachment of the cells. This step is important to avoid cell doublets.
- After addition, remove the trypsin directly with the same pipette tip.
- Incubate the cells for at least 5 min at 37 °C.
- Resuspend trypsinized cells with 1 mL of PBS containing 3% FCS and transfer the suspension in pre-labeled FACS tubes.
- Add DAPI (1 µg/mL) prior to the FACS analysis.
- Analyze the cells using a flow cytometer. For each sample, measure at least 10,000 living cells, which are negative for DAPI staining.
6. Data analysis
- Import the data to the flow cytometry analysis software and add samples by Click and Drag into the workspace.
- Double click on the imported file and create a graph plot. Choose FSC-A versus SSC-A by clicking on the x- and y-axis. Gate the main cell population by clicking on Polygon at the toolbar and framing the cell population (Figure 5). Name the chosen cell population as required (for example "single cells"). The "single cells" will appear as a new workspace.
- Proceed with gating "single cells" for DAPI negative (i.e., "living cells"). To identify living cells compare the cell population with and without DAPI staining. In both plots choose FSC-A versus pacific-blue-A.
- Click on the rectangle and choose the DAPI negative cell population, name the chosen cell population "living cells", and create a new dot-plot showing FITC (eGFP) versus PE (tdTomato) as described before.
- Add quadrants in "living cells" by choosing Quad from the toolbar. Gate the population by dragging the center of the quadrant to the edge of each population. The quadrants represent 1) eGFP-tdTOM-, 2) eGFP-tdTOM+, 3) eGFP+tdTOM-, 4) eGFP+tdTOM+.
NOTE: Due to the spectral overlay of emission spectra between FITC and PE, initial compensation is essential to distinguish between red and green signals and to achieve accurate results. - Determine mean fluorescence intensities (MFIs) by right clicking on the quadrant and choosing Add Statistics. Choose Mean and click OK. The mean MFI is now displayed below the population in the section "statistics". Quadrants 2 and 4 correspond to early expression, while quadrants 3 and 4 represent late expression.
- Add additional replicates in the same manner for statistical power.
- For data interpretation plot MFI values of early and late into a bar plot:
- To compare NCCR activities obtained from different donors or virus strains, add an archetypical control or the Dunlop strain16 and set its relative MFIs to 100%, and calculate the relative MFI values. Repeat with each replicate and determine mean and standard deviation.
- To evaluate an effect of potential compounds on the transcriptional activity, set the solvent-control to 100% and plot the MFI of the treated cells.
Representative Results
In this representative experiment, the BK-polyomavirus Non-Coding Control Region driven transcriptional activity was measured via flow cytometry. In addition, a mTOR inhibitor, which might be used to treat patients after BKPyV reactivation, was tested for its inhibition of the viral early gene expression. To this end, a dual fluorescence-reporter assay was used as published previously5. The overall workflow scheme of the experimental setup is illustrated in Figure 1. First, blood samples from immunocompromised renal transplanted patients were collected according to the guidelines of human research as approved by the institutional ethics committee. The blood was collected in EDTA-containing tubes and the phases were separated by centrifugation. Subsequently, 400 µL of plasma was used for isolation of the polyomavirus DNA. The non-coding control region (NCCR) was amplified using a nested-PCR protocol as illustrated in Figure 2A using the outer primer pair BKV-CR-1fw and BKV-CR-1rv, as well as, the inner primer pair BKV-CR-2fw-AgeI (Primer AgeI) and BKV-CR-2rv-SpeI (Primer SpeI)17, the latter of which harbors the restriction sites for AgeI and SpeI. The amplicon verification was performed via agarose gel electrophoresis as shown in Figure 2B. While the amplicons derived from archetypical NCCR sequences (wt) had a homogeneous size distribution in the gel, those derived from rearranged NCCRs with insertions (rrins) and deletions (rrdel) differed in their size (approx. 300-500 bp). Notably, due to rearrangements, the Dunlop NCCR reference sequence16 (DUN), which was included as a control, was larger than the NCCR obtained from archetypical viruses. Since the amplicons corresponded to the predicted sizes, the purified PCR products were sent for Sanger sequencing for later comparison with the sequences cloned into the reporter plasmid for further analysis.
For cloning of the amplicons, digestion was performed using the two restriction enzymes AgeI and SpeI. After cloning into the dual fluorescence reporter, which was performed using standard cloning procedure, selection of positive clones containing the NCCR was verified by AgeI and SpeI digestion (Figure 2C). Here, the small spacer region (NC, 128 bp) was replaced by the larger NCCR-fragments (approx. 300-500 bp) indicating positive clones. Plasmid DNA isolated from verified clones were sent for Sanger sequencing and compared with the sequences obtained from amplicon sequencing (not shown). Only the clones that match the amplicon sequence were chosen for further processing. As depicted in Figure 3, the sequencing results were compared with an archetypical NCCR sequence (JN192438). In this example, a deletion in the P-block and a substitution in O-block were detected.
To subsequently test the transcriptional activity of the cloned NCCR sequences in cell culture, HEK293T cells were transiently transfected with the respective reporter constructs (Figure 4A). As visualized in Figure 4B, the transfection efficiency and NCCR-activity were monitored via fluorescence microscopy. Red and green fluorescence corresponded to early and late BKPyV gene expression, respectively5. The cells expressed both fluorophores indicating efficient early and late promotor activity. As illustrated by the two examples, the first NCCR (NCCR1) displayed a strong late gene expression, while the second example (NCCR2) had a comparatively strong early but moderate late gene expression (Figure 4B). Thus, samples were prepared for the flow cytometry measurement. As described in protocol section 6, DAPI negative cells were gated (Figure 5A, lower panel) and a dot plot was created showing eGFP versus tdTomato. Samples that were negative (Figure 5B), as well as those positive for tdTomato (Figure 5C) or eGFP (Figure 5D) only, were included into the analysis. Note, initial compensation was performed, which is essential to distinguish red and green signals and to achieve accurate results18. Mean fluorescence intensities were derived from each testing clone. Here, tdTomato positive cells corresponded to early expression while eGFP positive cells represented late expression. In this example the treatment with the dual mTOR inhibitor INK128 significantly reduced tdTomato MFI (Figure 5E-F), which means that early expression was inhibited.
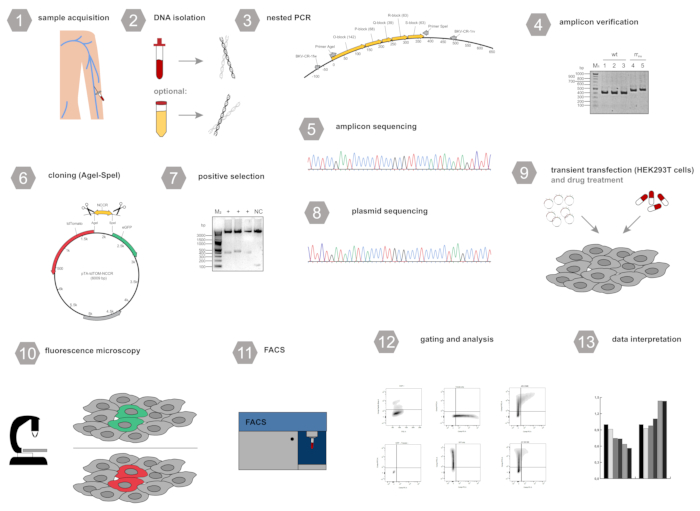
Figure 1: Workflow scheme for the experimental setup.1) Collection of blood (alternatively urine) samples, 2) Isolation of polyomavirus DNA 3) Amplification of the non-coding control region (NCCR) using a nested-PCR protocol, 4) Agarose gel electrophoresis and amplicon verification, 5) Sanger sequencing of the PCR amplicon, 6) Cloning of the NCCR into the dual fluorescence reporter using AgeI and SpeI, 7) Selection of positive clones, 8) Sanger sequencing of the plasmid DNA, 9) Transient transfection of HEK293T cells with the reporter plasmid and addition of compounds to be tested, 10) Fluorescence microscopy to verify efficient transfection, 11) Flow cytometry analysis, 12) Gating and analysis of eGFP tdTomato expression, 13) Data interpretation. Please click here to view a larger version of this figure.
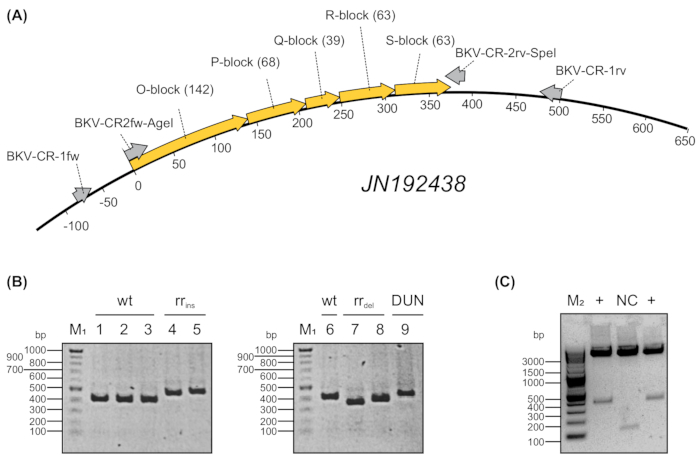
Figure 2: Nested PCR and representative analysis of patient derived NCCR-amplicons.A) DNA derived from immunocompromised renal transplantedpatients was subjected to semi-nested PCR amplification using oligonucleotide primers linked with restriction sites AgeI and SpeI. Primer names and length (base pairs) of archetypical O, P, Q, R, and S-blocks from BKPyV strain JN192438 are indicated in brackets. B) Amplicons were analyzed by electrophoresis in 1.5% agarose gel and visualized using DNA staining. M1 = 100 bp ladder, wt = wildtype (archetypical), rr = rearranged, ins = insertions, del= deletions, DUN = Dunlop Strain. C) Plasmid digestion with AgeI and SpeI. Digested fragments were analyzed by electrophoresis in 1.5% agarose gel and visualized using DNA staining. M1 = 100 bp ladder, M2 = 2-log ladder. NC = negative control (spacer). + = positive clone. Please click here to view a larger version of this figure.
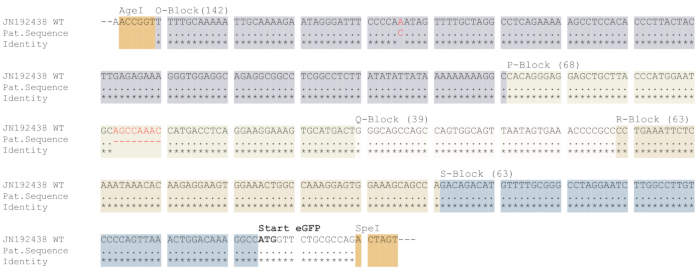
Figure 3: Cloning and validation of the NCCR into the dual fluorescence reporter. Representative results of plasmid sequencing. Amplicons were purified using a PCR purification kit and subjected to Sanger sequencing. The respective sequences were aligned to the NCCR sequence derived from archetypical BKPyV strain JN192438. Deletions and substitutions are marked in red. The AgeI and SpeI restriction sites, translational start of eGFP open reading frame (printed in bold) as well as the NCCR blocks O-S are indicated. The respective blocks and the restriction sites are highlighted in color. The length of each NCCR block in base pairs are printed in brackets. Please click here to view a larger version of this figure.
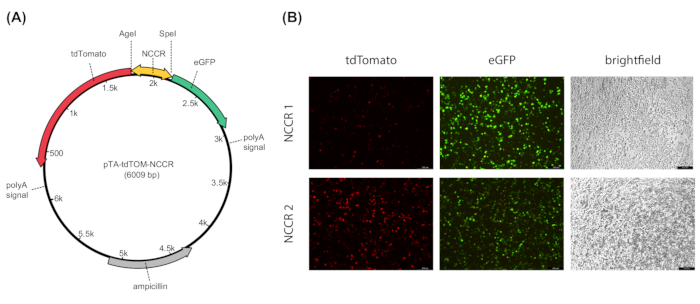
Figure 4: Representative fluorescence microscopy analysis of early and late NCCR promoter activity.A) Gene map of the dual fluorescence reporter under the control of the bidirectional promoter. As described previously5 the plasmid pTA-tdTOM-NCCR-eGFP (6,009 bp) harbors the SV40 late polyadenylation signal downstream of each expression cassette to ensure comparable and efficient processing of both transcripts for tdTomato (early expression) and eGFP (late expression), respectively. NCCR is flanked by AgeI and SpeI. The vector contains an ampicillin resistance cassette for selection during cloning procedure. B) HEK293T cells were transfected with dual-fluorescence reporter constructs each under the control of patient derived NCCR sequences (NCCR1-2). Cells were subjected to brightfield and fluorescence microscopy analysis 72 h post transfection. Filter settings of the microscope were used in the following excitation ranges: green for tdTomato (515-560 nm) and blue for eGFP (450-490 nm). The scale bar (200 µm) is indicated in the bottom right of each photography. Please click here to view a larger version of this figure.
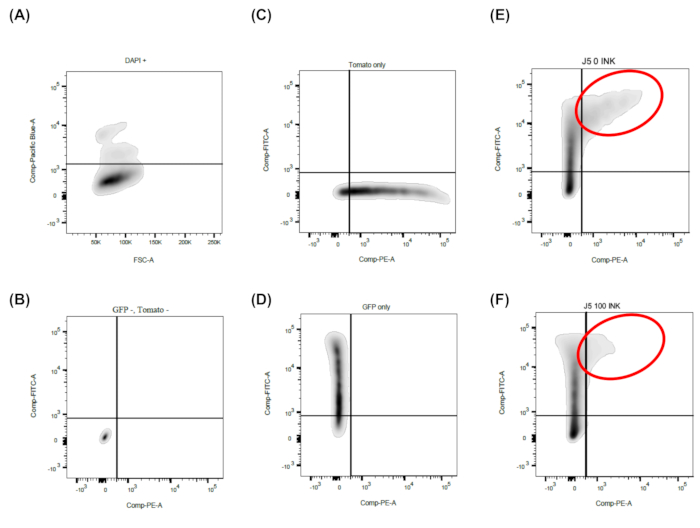
Figure 5: Representative FACS analysis. Shown is the simple gating strategy required for this method. Two-parameter density or dot plots are used. A) First, FSC is plotted against Pacific Blue, while only DAPI negative (i.e., living cells) are further processed. B-F) FITC (eGFP) versus PE (tdTomato) are plotted. C-D) Add exclusively green and red fluorescent cells to adjust the compensation on the flow cytometer. E-F) In this example, the mTOR inhibitors INK128 and significantly reduces the tdTomato MFI, which corresponds to a reduction of the BKPyV early gene expression. Please click here to view a larger version of this figure.
| Primer name | Sequence (5' → 3') |
| BK-CR-fwd1 | CCCAGGCAGCTCTTTCAAGGC |
| BK-CR-rev1 | CCTCTAACAAAATTCCAGCAAAAGC |
| BKV-CR-2-fw-AgeI | AAAAAAACCGGTTTTTGCAAAAATTGCAAAAGAATAGG |
| BKV-CR-2-rv-SpeI | TTTTTTACTAGTCTGGCGCAGAACCATGGCCTT |
| EGFP-N | CGTCGCCGTCCAGCTCGACCAG |
Table 1: Oligonucleotides used in this protocol. The underlined bases indicate the restriction enzyme sites for AgeI and SpeI, respectively.
| Component | Volume/reaction (µl) | final concentration |
| RNase-free water | 32.8 µL | – |
| 10x PCR Buffer | 5 µL | 1x |
| dNTPs (10 mM of each) | 1 µL | 0.2 mM of each dNTP |
| Fwd-primer (10 µM) | 3 µL | 0.3 µM |
| Rev-primer (10 µM) | 3 µL | 0.3 µM |
| Taq Polymerase | 0.2 µL | 2 unit/reaction |
| Template DNA | 5 µL | <0.5 µg/50 µL reaction |
Table 2: Master mix preparation protocol. The instruction applies to both pre- and nested PCR reactions as described in steps 2.2 and 2.7, respectively.
| Temperature | Duration | PCR-step | cycles |
| 95°C | 5 min | Initial Denaturation | |
| 95°C | 30 sec | Denaturation | |
| 55°C | 34 sec | Annealing | x35 |
| 72°C | 1 min | Extension | |
| 72°C | 10 min | Final extension | |
| 4°C | ∞ | Cooling |
Table 3: PCR program used for NCCR amplification. The instruction applies to both pre- and nested PCR reactions.
| Reagent | Volume/reaction (µl) | final concentration |
| DNase-free H20 | 6 | to 20 µl |
| 10x Buffer | 2 | 1x |
| AgeI | 1 | 10 units |
| SpeI | 1 | 10 units |
| purified PCR-amplicon | 10 | variable |
Table 4: Amplicon digestion protocol. The instruction applies for the simultaneous digestion of the amplicon DNA with two restriction enzymes described in step 3.1.
| Reagent | Volume/reaction (µl) | final concentration |
| DNase-free H20 | 14.5 | to 20 µl |
| 10x Buffer | 2 | 1x |
| AgeI | 1 | 10 units |
| SpeI | 1 | 10 units |
| Plasmid backbone (1 µg/µL) | 1.5 | 1.5 µg |
Table 5: Plasmid digestion protocol. The instruction applies for the simultaneous digestion of the plasmid DNA backbone with two restriction enzymes described in step 3.3.
| Reagent | Volume/reaction (µl) | final concentration |
| DNase-free H20 | 7 | to 20 µl |
| T4 DNA ligase | 1 | 20 |
| 10x T4 DNA ligase buffer | 2 | 1x |
| Purified Plasmid backbone Low melt diluted ½ from step 3.5 |
2 | variable |
| Digested and purified PCR-amplicon from step 2.7 | 8 | variable |
Table 6: Plasmid ligation protocol. The instruction applies for the ligation of the plasmid DNA backbone with the digested PCR amplicon DNA described in step 3.7.
Discussion
In this article, a commonly used method is presented that allows for the analysis of the BKPyV non-coding control-region (NCCR) driven early and late promoter activity. The NCCR activity can be measured simultaneously and does not need lysis of the transfected cells. Furthermore, a relatively large number of cells can be analyzed and the co-transfection of additional markers for normalization of the fluorescence values is not necessary.
A critical part of this method is that the cloned NCCR should contain exactly the same sequences as identified by the amplicon sequencing, which corresponds to the majority variants present in the patient plasma. To achieve accurate results and to minimize PCR-prone errors it is highly recommended to use a polymerase with proof reading activity. Alternatively, sequences can be ordered by commercial gene synthesis services. The designated NCCR sequences can be ordered including flanking AgeI and SpeI restriction sites for direct cloning. In this case, digest the plasmid parallel to the backbone plasmid and replace the spacer insert with the corresponding NCCR fragment from synthesized construct. DNA can be alternatively isolated from urine samples. However, since BKPyV is also secreted into urine by immunocompetent individuals and not necessarily reflect active replication, the clinical relevance is limited. Ideally, the DNA can also be isolated from renal biopsies in which previously PVAN could be histologically detected (compare with Korth et al., 201919).
By measuring the mean fluorescence intensity of green and red fluorescent cells, this method allows to quantify NCCR-derived fluorescence/activity without the bias of transfection efficiency and anti-proliferative effects of the tested agents (e.g., dual mTOR inhibitors INK128 or PP242)5.
Another application of the reporter system is the possibility of analyzing the consequences of insertions, deletions, or duplications in the NCCR found in clinical isolates on the early and late promotor activity. In previous studies, immunosuppressive agents have been studied to modulate NCCR activity; however, no mutations or NCCR rearrangements were found that affect the susceptibility. However, mutations may affect the response of new substances that might be approved in the near future.
This might be relevant since in contrast to the coding regions large T antigen or VP1 capsid protein, the NCCR as the name implies represents a noncoding region and thus does not underlie selective pressure at the amino acid level.
In this protocol, the selection of positive clones containing the NCCR is verified by AgeI and SpeI digestion. However, a colony PCR might represent an alternative approach to quickly screen for NCCR inserts directly from E. coli plated on agar plates. For this approach, use primer pair BKV-CR-2-fw-AgeI and EGFP-N (Table 1). After transfection, cells are checked for red and green fluorescence using fluorescence microscopy (Figure 4). Alternatively, cells can be fixed using 3% PFA for 20 min at 22 °C. In this case, wash with PBS, add 0.2% non-ionic detergent in PBS and incubate for 10 min. Fixed cells can then be stained with DAPI in PBS (1 µg/mL) for 10 min at 22 °C and subjected to fluorescence microscopy analysis and visualized with UV light (340-380 nm).
Since both fluorophores harbor the same polyadenylation signals, an effect of polyadenylation efficiency can be out ruled. However, in comparison to eGFP, tdTomato is a dimeric protein which must be correspondingly transcribed into a longer mRNA (coding sequence: 745 versus 1431 bp, respectively). However, since the activity of the promoters from the cells treated with interfering substances are always compared relative to untreated (DMSO) cells, this difference plays little role. Due to the presence of the origin of replication, the transfection of the reporter plasmid in cells stably expressing the SV40PyV large T antigen allows the transfer of plasmids to the daughter cells. It has been shown that the SV40 derived large T antigen can transactivate SV40 and BKPyV NCCR and viral DNA replication20. However, since a large T antigen is also involved in the transition from the early to the late phase of infection, an artificial situation is given. Using this assay, it must be considered that the most replication limiting step is the early expression leading to the expression of large T antigen. In order to map the complete replication cycle the early and late mRNAs should be measured in infected cells by qRT-PCR as described elsewhere5,12.
A comparable method has already been used by Swiss and German groups to analyze archetypical and rearranged BKPyV NCCR-derived promotor activities, albeit with independently cloned vector systems5,9. Gosert and colleagues from Basel used a similar method to determine the NCCR promoter activity of BKPyV and JCPyV strains9,10,11. Both methods used eGFP for green fluorescence; however, the Basel group used RFP while in the current protocol tdTomato was used for red fluorescence. The advantage of tdTomato over RFP is the much higher photostability; however, the dimeric protein is encoded by a sequence that is twice as long as RFP21. Furthermore, the half-lives of the proteins might be different, resulting in unequal concentration 72 h post transfection. However, this issue might only be relevant when directly comparing both fluorescence intensities. Gosert et al. used BamHI and NotI as restriction sites while in the current protocol AgeI and SpeI are flanking the NCCR sequence. Using both methods, the reference DUNLOP strain16 yielded comparable fluorescence patterns with strong early but weak late promoter activity9,10. In further agreement, NCCR-rearrangements with insertions in the P-region resulted in a significant increase in early and late promoter activity when compared to wt-NCCR5,9.
Although it has been shown that the SV40 derived large T antigen can transactivate SV40 and BKPyV derived NCCRs20, it might be interesting to use human cells that stably express the large T antigen of the BKPyV and not the prototype virus SV40 in future studies. However, in this case the cell line has to be easy to transfect (e.g., HEK293).
Since the NCCR promoters of polyomaviruses are very similar, this method can also be used (with small adaptation in the primer sequences) to investigate promoter activity and the influence of potent antiviral agents on the activity of JC-polyomavirus (JCPyV) derived non coding control regions11. Since JCPyV is the causative agent of the progressive multifocal leukoencephalopathy (PML), a rare central nervous system disease that can rarely occur in patients with immunodeficiencies resulting in severe neuropathology, there is also a pressing need to find direct acting antiviral substances to treat immune compromised patients. Interestingly, rearrangements are also found in JC NCCRs11. Since JCPyV has pronounced neurotropism and thus large quantities of viruses are found in liquor, sample acquisition has to be adjusted to liquor-derived DNA. However, subsequent steps can be easily adapted.
Declarações
The authors have nothing to disclose.
Acknowledgements
The authors thank Barbara Bleekmann for excellent technical assistance. These studies were supported by the IFORES-program of the University of Duisburg-Essen Medical School and the RIMUR-program of the University Alliance Ruhr and Mercator Research Center Ruhr (MERCUR). The authors thank the Jürgen-Manchot-Stiftung for the doctoral fellowship of Helene Sertznig and constant support. The collection and use of patient material has been approved by the ethics committee of the medical faculty of the University Duisburg-Essen (14-6028-BO).
Materials
| 100 bp ladder | NEB | N3231 | Any ladder with a range up to 1 kb can be substituted |
| 2-log ladder | NEB | N0550 | Any ladder with a range up to 10 kb can be substituted |
| Agar-Agar, Kobe I | Carl Roth | 5210.3 | |
| AgeI-HF | NEB | R3552 | |
| Ampicillin Natriumsalz Cellpure | Carl Roth | HP62.1 | |
| Aqua ad iniectabilia | Bbraun | 2351744 | can be substituted by any manufacturer |
| BD FACSCanto™ II | BD Biosciences | ||
| BD FACSDiva | BD Biosciences | ||
| DAPI | Sigma | 10236276001 | |
| DFC450C camera module | Leica | Any camera can be used | |
| DMEM | Gibco | 41966-029 | can be substituted by any manufacturer |
| DMIL LED microscope | Leica | Any fluorescence microscope can be used | |
| DNA Blood Mini Kit | Qiagen | 51104 | can be substituted by any manufacturer |
| E.coli DH5alpha Competent Cells | Thermo Scientific | 18258012 | |
| FBS Superior | MerckMillipore | 50615 | Any FBS can be used |
| FlowJo v10.5.3 | FlowJo, LLC | Any flow cytometry software FBS can be used | |
| Gel Loading Dye, Purple (6X) | NEB | B7024 | Any 6x loading dye can be substituted |
| HEK293T cells | ATCC | 11268 | These cells constitutively express the simian virus 40 (SV40) large T antigen, and clone 17 was selected specifically for its high transfectability. |
| HERAcell® 240i CO2 Incubator | Thermo Scientific | can be substituted by any manufacturer | |
| HotStar PCR kit | Qiagen | 203203 | |
| Intas Gel documentation system | Intas | Any visualisation system for stained DNA containing agarose gels can be used | |
| Low Melt Agarose | Biozym | 850081 | can be substituted by any manufacturer |
| Opti-MEM | Invitrogen | 31985070 | can be substituted by any manufacturer |
| pBKV (34-2) | ATCC | 45025 | Plasmid harboring the full-length genome of BKPyV strain Dunlop; was used as a positive control; DNA Seq. Acc.: KP412983 |
| PBS | Gibco | 14190-136 | |
| PCR Cycler MJ Mini 48-Well Personal Cycler | Bio-Rad | discontinued product | Any thermocycler can be used |
| PCR Nucleotide Mix, 10 mM | Promega | #C1145 | can be substituted by any manufacturer |
| PCR1 and PCR2 Primers | metabion | not applicable | Desalted. Dilute to (10 µM) with PCR grade water |
| PenStrep (100x) | Gibco | 15140-122 | can be substituted by any manufacturer |
| Roti®-GelStain | Carl Roth | 3865 | A fluorescence based stain for measuring dsDNA concentration |
| SpeI-HF | NEB | R3133 | |
| T4-DNA Ligase | NEB | M0202 | |
| TransIT LT1 | Mirus | MIR2300 | |
| Trypsin 0.05% – EDTA | Gibco | 25300-054 | can be substituted by any manufacturer |
| ZymoPURE II Plasmid Kit | Zymo | D4201 | can be substituted by any manufacturer |
Referências
- Drachenberg, C. B., et al. Histological patterns of polyomavirus nephropathy: correlation with graft outcome and viral load. American Journal of Transplant. 4 (12), 2082-2092 (2004).
- Korth, J., et al. Impact of low-level BK polyomavirus viremia on intermediate-term renal allograft function. Transplant Infectious Disease. 20 (1), (2018).
- Hirsch, H. H., et al. European perspective on human polyomavirus infection, replication and disease in solid organ transplantation. Clinical Microbiology and Infection. 20 Suppl 7, 74-88 (2014).
- Masutani, K., et al. The Banff 2009 Working Proposal for polyomavirus nephropathy: a critical evaluation of its utility as a determinant of clinical outcome. American Journal of Transplantation. 12 (4), 907-918 (2012).
- Korth, J., et al. Impact of immune suppressive agents on the BK-Polyomavirus non coding control region. Antiviral Research. 159, 68-76 (2018).
- Hirsch, H. H., Yakhontova, K., Lu, M., Manzetti, J. BK Polyomavirus Replication in Renal Tubular Epithelial Cells Is Inhibited by Sirolimus, but Activated by Tacrolimus Through a Pathway Involving FKBP-12. Amerian Journal of Transplantation. 16 (3), 821-832 (2016).
- Sanchez Fructuoso, A. I., et al. Mammalian target of rapamycin signal inhibitors could play a role in the treatment of BK polyomavirus nephritis in renal allograft recipients. Transplant Infectious Disease. 13 (6), 584-591 (2011).
- Moens, U., Johansen, T., Johnsen, J. I., Seternes, O. M., Traavik, T. Noncoding control region of naturally occurring BK virus variants: sequence comparison and functional analysis. Virus Genes. 10 (3), 261-275 (1995).
- Gosert, R., et al. Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. Journal of Experimental Medicine. 205 (4), 841-852 (2008).
- Bethge, T., et al. Sp1 sites in the noncoding control region of BK polyomavirus are key regulators of bidirectional viral early and late gene expression. Journal of Virology. 89 (6), 3396-3411 (2015).
- Gosert, R., Kardas, P., Major, E. O., Hirsch, H. H. Rearranged JC virus noncoding control regions found in progressive multifocal leukoencephalopathy patient samples increase virus early gene expression and replication rate. Journal of Virology. 84 (20), 10448-10456 (2010).
- Bernhoff, E., Gutteberg, T. J., Sandvik, K., Hirsch, H. H., Rinaldo, C. H. Cidofovir inhibits polyomavirus BK replication in human renal tubular cells downstream of viral early gene expression. American Journal of Transplantation. 8 (7), 1413-1422 (2008).
- Widera, M., et al. HIV-1 persistent viremia is frequently followed by episodes of low-level viremia. Medical Microbiology Immunology. , (2017).
- Zhang, S., Cahalan, M. D. Purifying plasmid DNA from bacterial colonies using the QIAGEN Miniprep Kit. Journal of Visualized Experiment. (6), 247 (2007).
- Drew, R. J., Walsh, A., Laoi, B. N., Crowley, B. Phylogenetic analysis of the complete genome of 11 BKV isolates obtained from allogenic stem cell transplant recipients in Ireland. Journal of Medical Virology. 84 (7), 1037-1048 (2012).
- Dorries, K., Vogel, E., Gunther, S., Czub, S. Infection of human polyomaviruses JC and BK in peripheral blood leukocytes from immunocompetent individuals. Virology. 198 (1), 59-70 (1994).
- Teutsch, K., et al. Early identification of renal transplant recipients with high risk of polyomavirus-associated nephropathy. Medical Microbiology Immunology. 204 (6), 657-664 (2015).
- Basu, S., Campbell, H. M., Dittel, B. N., Ray, A. Purification of specific cell population by fluorescence activated cell sorting (FACS). Journal of Visualized Experiment. (41), (2010).
- Korth, J., et al. The detection of BKPyV genotypes II and IV after renal transplantation as a simple tool for risk assessment for PyVAN and transplant outcome already at early stages of BKPyV reactivation. Journal of Clinical Virology. 113, 14-19 (2019).
- Caputo, A., Barbanti-Brodano, G., Wang, E., Ricciardi, R. P. Transactivation of BKV and SV40 early promoters by BKV and SV40 T-antigens. Virology. 152 (2), 459-465 (1986).
- Shaner, N. C., Steinbach, P. A., Tsien, R. Y. A guide to choosing fluorescent proteins. Nature Methods. 2 (12), 905-909 (2005).

