Mapping Regional Homogeneity and Functional Connectivity of the Visual Cortex in Resting-State fMRI
Summary
We present a protocol for analyzing functional magnetic resonance imaging data to investigate spontaneous neural activity alterations in retinitis pigmentosa patient using a combined regional homogeneity and functional connectivity method.
Abstract
A combined regional homogeneity (ReHo) and functional connectivity (FC) method, a type of noninvasive functional magnetic resonance imaging (fMRI) method, has been used to evaluate synchronous neuronal activity changes in retinitis pigmentosa (RP). The purpose of this study is to describe our method for analysis of intra- and interregional synchronizations of changes in neuronal activity in RP patients. The advantages of the combined ReHo and FC method are that it is both noninvasive and sufficiently sensitive to investigate changes in cerebral synchronous neuronal activity changes in vivo. Here, 16 RP patients and 14 healthy controls closely matched in age, sex, and education underwent resting-state fMRI scans. Two sample t-tests were conducted to compare ReHo and FC across groups. Our results showed that visual network disconnection and reorganization of the retino-thalamocortical pathway and dorsal visual stream occurred in the RP patients. Here, we describe the details of this method, its use, and the impact of its key parameters in a step-by-step manner.
Introduction
Functional magnetic resonance imaging (fMRI) is a noninvasive method that can be used to investigate alterations in brain function and structure in vivo. Regional homogeneity (ReHo) and functional connectivity (FC) are often used to assess intra-and interregional synchronizations of brain activity. ReHo, a resting-state fMRI methodology, is used to calculate similarity between the time series of a given voxel and its nearest neighbors, which reflects the local synchronization of brain activities1. FC is used to investigate the similarity between spatially remote regional time series2.
fMRI technology can offer an objective assessment of visual function in the context of eye disease management. Here, we present a methodological protocol that combines ReHo and FC methods to share this experience and support the dissemination of our expertise. In the present work, we used the ReHo and FC protocol in retinitis pigmentosa (RP) subjects and healthy controls (HCs) to elaborate the details of the procedure. RP is a serious hereditary eye disease characterized by impaired night vision and the progressive loss of vision3,4. Genetic mutation is the main risk factor for RP. The death of rod and cone photoreceptor cells leads to the loss of peripheral vision and finally blindness in RP patients. Previous neuroimaging studies have shown structural and functional abnormalities in the visual cortex and visual pathway of RP patients5,6,7. Moreover, diffusion tensor imaging was used to investigate the integrity of white matter fiber bundles. RP patients showed significantly higher apparent diffusion coefficient, principal eigenvalue, and orthogonal eigenvalue, as well as significantly lower fractional anisotropy in the optic nerves, relative to HCs8.
Here, our aim was the exploration of intra- and interregional synchronizations of neuronal activity. We investigated whether the mean ReHo values and mean FC values were correlated with clinical variables in RP patients. Our method might enable researchers to obtain important insights into the neural mechanism of peripheral vision loss in RP patients.
Protocol
The research protocol was approved by the medical ethics committee of the Renmin Hospital of Wuhan University. All participants completed a written consent form.
1. Participant classification and screening
- Enroll RP subjects and HCs closely matched in age, gender, and education.
- Ensure that all participants meet the following criteria: 1) able to be scanned with an MRI scanner (e.g., no cardiac pacemakers or implanted metal devices); 2) no claustrophobia; 3) no heart disease, hypertension, or cerebral diseases.
2. Acquisition of fMRI data
NOTE: A 3 T MRI scanner with eight-channel head coil is used in this protocol.
- Ask each participant to remove metal objects before entering the MRI scanner room after a final safety check.
- Instruct the participant to lie down on the bed and ensure that the orbitomeatal line is perpendicular to the bed. Then place foam pads in the bilateral temporal region of the head to prevent head movement and provide earplugs to reduce the noise of the scanner.
- Instruct the participant to lie at rest, to keep his/her eyes closed without falling asleep, and not think of anything in particular during scanning.
- Adjust the participant’s head position through the positioning light. Make sure that the axis positioning cursor is parallel to the lateral canthus and the sagittal positioning cursor coincides with the midline of the face. Next, move the bed to make the axis positioning cursor stay 2 cm above or below the participant’s eyebrows.
- Notify the participant of the start of the scanning session. Using the scanning console, start the structural localizer scanning to determine the position of the participants’ head in the scanner and allow planning for subsequent structural and functional scans.
- Perform fMRI with the following sequences and parameters.
- Perform three-dimensional brain volume imaging (3D-BRAVO) MRI with the following parameters: repetition time (TR)/echo time (TE) = 8.5 ms/3.3 ms; thickness = 1.0 mm; no intersection gap; acquisition matrix = 256 x 256; field of view = 240 x 240 mm2; and flip angle = 12°.
- Obtain functional images using gradient echo-planar imaging blood oxygenation level-dependent (EPI-BOLD) imaging with the following parameters: TR/TE = 2,000 ms/25 ms; thickness = 3.0 mm; gap = 1.2 mm; acquisition matrix = 256 x 256; field of view = 240 x 240 mm2; voxel size = 3.6 x 3.6 x 3.6 mm3; and 35 axial slices.
- Keep an eye on the condition of the participant during the duration of the scan, instruct them to move as little as possible, and stop scanning if the participant has any discomfort.
- Remove the participant from the scanner and ask the participant to sit up carefully at the end of the experiment.
3. Data preprocessing and software preparation
NOTE: The functional images analyzed in this protocol are preprocessed by SPM8 and the toolbox for Data Processing & Analysis for Brain Imaging (DPABI, http://rfmri.org/dpabi)9 based on MATLAB 2013a. Perform the following preprocessing steps separately for each fMRI session.
- Open the DPABI software in the MATLAB terminal by clicking dpabi, then choose DPARSF 4.3 Advanced Edition and import the folder "FunRaw" (Figure 1).
NOTE: The FunRaw folder contains the DICOM file for each participant. - Click FunRaw to import the fMRI scan files into DPABI with a consistent numbering scheme (e.g., "sub0001", "sub0002", etc.). Select the working directory and the initial EPI and T1 directories and continue to select all desired parameters in steps 3.3–3.9 before clicking Run in section 4.
- Type the parameters: Timing points = 240 and TR = 2 (Figure 2). Select the EPI DICOM to NIFTI to convert the functional images from DICOM to NIFTI format and remove the first 10 volumes of each function images.
- Check the boxes for Slice Timing and Realign in the DPABI software to correct the remaining 230 volumes of functional blood oxygenation level-dependent images for slice timing effects and head motion corrected.
NOTE: For head motion, the data of participants with head movement >2 mm or rotation >2° during scanning should be excluded. For slice order, the sequence of numbers in the vector is the acquisition time order of these layers. The selected slice number is 40, slice order is [1:2:39,2:2,40], and the reference slice is 39. - Select Normalize by DARTEL with the DPABI software.
NOTE: By selecting this option, the software will automatically perform spatial normalization using individual T1-weighted structural images registered to the mean fMRI data. The resulting aligned T1-weighted images are segmented using the DARTEL toolbox for improved spatial precision during normalization of fMRI data. Normalized data (in Montreal Neurological Institute [MNI] 152 space) are resliced at a resolution of 3 x 3 x 3 mm3. - Remove the linear trend by selecting Detrend in the DPABI software.
- Check the box for Nuisance Covariates Regression and select the following parameters: head motion model, white matter signal, global mean signal, and cerebrospinal fluid signal.
- Check the box for Scrubbing to remove the bad time points due to head motion in the DPABI software.
- Retain signals between 0.01–0.08 Hz by checking the box Filter [0.01-0.08] in the DPABI software to remove high-frequency physiological noise and low-frequency drift.
NOTE: After the data preparation, ReHo and FC analysis can be performed.
4. ReHo and FC analysis
- For the ReHo computation, open the DAPABI software through MATLAB and select 27 voxels in the cluster. Left click ReHo and smooth [6*6*6], then select Run.
NOTE: A Kendall's coefficient of concordance is assigned to a given voxel by calculating the Kendall’s coefficient of concordance of time series of 27 voxels and their nearest neighbors. To reduce the influence of individual variations on statistical comparisons between groups, ReHo maps of each voxel are z-transformed using Fisher’s r-to-z transformation. The remaining z ReHo maps are spatially smoothed using a Gaussian kernel of 6*6*6 full width at half maximum. - For the FC computation, open the DAPABI software through MATLAB and define the altered ReHo brain regions between both groups as regions of interest (ROIs). Click on Functional Connectivity and define the ROI (centering at x = 0, y = -69, and z = -3 with radius = 10 mm), then select Run.
NOTE: Correlation analysis of the time course for each participant is performed between the spherical seed region and whole brain voxels. All FC maps are z-transformed by Fisher’s r-to-z transformation to reduce the influence of individual variations on statistical comparison between groups. The radius of the ROIs around the coordinates should be 10 mm (X = 0, Y = -69, Z = -3).
5. Statistical analysis
- Find the folders named ReHo and FC after processing the relevant file data. Sort the files of zReHo.nii and zFC.nii, classifying them into four subfolders: "RP-group-ReHo", "HC-group-ReHo", "RP-group-FC", and "HC-group-FC".
- Open DPABI through MATLAB to perform a one sample t-test.
- Left click Statistical Analysis, then click one-Sample t-test. Name the output result "one-sample-t-test-RP" and set the output directory.
- Left click Add Group Images and open the "RP-group-ReHo" subfolder.
- In the Mask File option, left click to open the "BrainMask-05-61*73*61" subfile in the "mask" folder.
- Select Compute to run the program. Perform this same procedure for the "one-sample-t-test-HC" group.
NOTE: The one-sample t-test is used to analyze and display mean ReHo maps of each group in DPABI software.
- Open DPABI through MATLAB to perform a two sample t-test.
- Left click Statistical Analysis, then select two-sample t-test. Name the output result "two-sample-t-test-ReHo" and set the output directory.
- Left click Add Group Images and open the "RP-group-ReHo" and "HC-group-ReHo" subfolders.
- In the Mask File option, left click to open the "BrainMask-05-61*73*61" subfile in the "mask" folder.
- Select Compute to run the program. Perform this same procedure for the "two-sample-t-test-FC". Click Statistical Analysis and Gaussian random field (GRF) correction [two-tailed, voxel-level (0.01) and voxel-level (0.05)] and then click Run.
NOTE: The differences between groups of zReHo maps and zFC maps are compared by two sample t-tests. GRF is used to correct for multiple comparisons and regressed covariates of age and sex with DPABI software.
- Use BrainNet Viewer software (https://www.nitrc.org/projects/bnv/) to show the results.
- Open BrainNet through MATLAB and click on Load file. For surface files, click Explorar and select BrainMesh-ICBM152-smoothed.nv, then click Ok; for volume files, select spm-T.nii (this includes ReHo and FC results), then click Ok.
- Use statistical software to process the data obtained from the previous step.
NOTE: The chi-squared test is used for sex comparisons, while independent-samples t-tests are used for other clinical variables. Continuous variables are represented by means and standard deviations. - Conduct Pearson correlation coefficient analysis to identify the relationships between zReHo values and zFC values of different brain regions and visual measurements data by using statistical software.
- Obtain ROI signals of zReHo values and zFC values in each participant by DPABI software. Click ROI signals extractor and Add directory with ROI mask.nii file.
NOTE: P values of <0.05 should be considered statistically significant.
- Obtain ROI signals of zReHo values and zFC values in each participant by DPABI software. Click ROI signals extractor and Add directory with ROI mask.nii file.
Representative Results
In our study, 16 RP individuals and 14 healthy controls closely matched in age, sex, and education underwent resting-state fMRI scans. ReHo and FC methods were used to explore the intra-and intersynchronous neuronal activity in RP individuals. Significant differences in BCVA were observed between the right eye (P < 0.001) and the left eye (P < 0.001), but the difference in gender, age, or weight between the groups was not significant.
The RP and HCs show similar spatial distribution in the ReHo maps. However, the ReHo value of the visual area in RP patients was significantly lower than that in the control group (Figure 3A,B). Compared with HCs, the ReHo values of the RP individuals were significantly lower in the bilateral LGG/CPL (BA 17,18) compared to the HCs (Figure 4 and Table 1) (Two-tailed, voxel-level P < 0.01, GRF correction, cluster-level P < 0.05).
Compared with the HCs group, the RP group showed increased FC between the bilateral LGG/CPL and bilateral thalamus and decreased FC between the bilateral LGG/CPL and left postcentral (Figure 5 and Table 2).
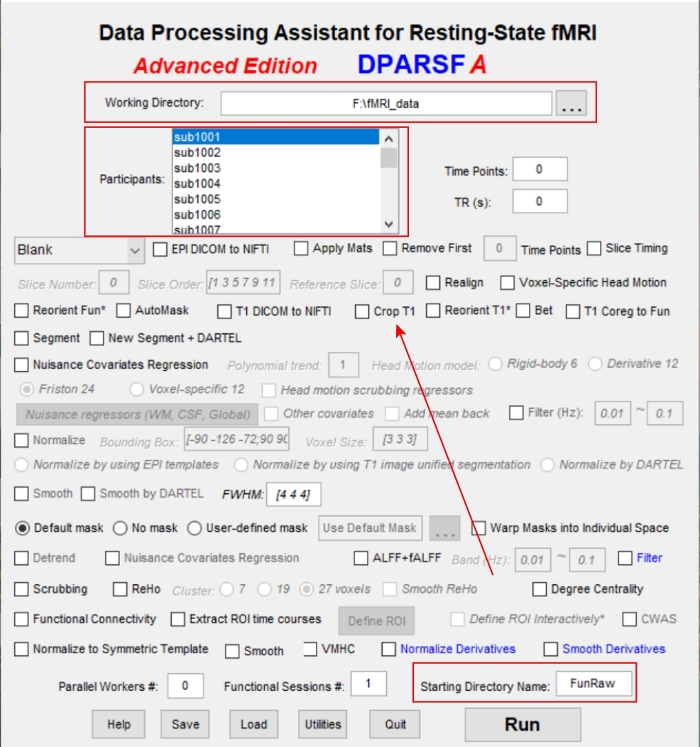
Figure 1: The operation interface of the DEPASFA toolbox. Please click here to view a larger version of this figure.
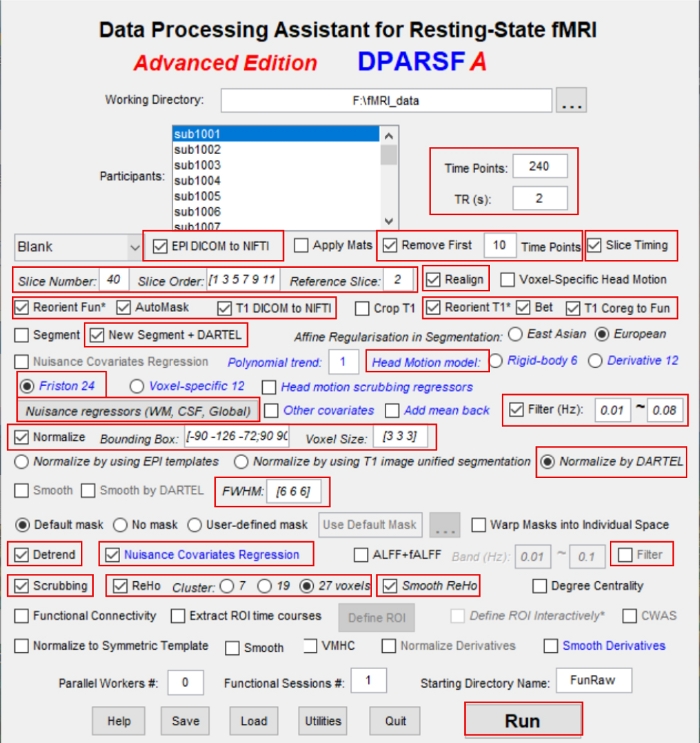
Figure 2: The operation interface of the DEPASFA toolbox with parameters entered. Please click here to view a larger version of this figure.
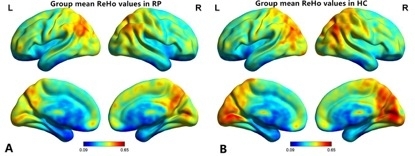
Figure 3: The distribution pattern of ReHo values in the RP and HC participants in the typical frequency band (0.01–0.08 Hz). Within-group means ReHo maps within the RP participants (A) and the HCs (B). ReHo = regional homogeneity; RP = retinitis pigmentosa; HC = health control; L = left hemisphere; R = right hemisphere. Please click here to view a larger version of this figure.
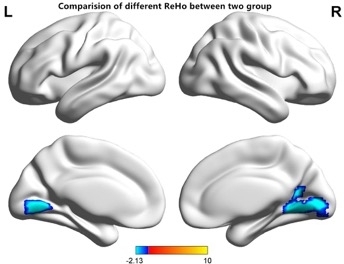
Figure 4: Comparisons of the ReHo values between the RP and HCs participants. There were significant regional differences in spontaneous activities between the two groups. ReHo values of RP participants were significantly lower in the bilateral LGG/CPL (BA 17,18) compared to those of HCs. The blue areas indicate lower ReHo values (two-tailed, voxel-level P < 0.01, GRF correction, cluster-level P < 0.05). GRF = Gaussian random field; LGG = lingual gyrus; CPL = cerebellum posterior lobe. Please click here to view a larger version of this figure.
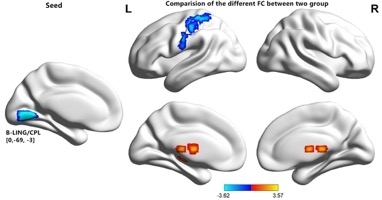
Figure 5: Comparisons of seed-based FC of the altered ReHo between the RP and HC groups. There were significant differences in seed-based FC activities between the two groups. The color-bars indicate the T-values. FC = functional connectivity; L = left hemisphere; R = right hemisphere; GRF = Gaussian random field; LGG = lingual gyrus; CPL = cerebellum posterior lobe. Please click here to view a larger version of this figure.
| Brain regions | BA | T-Peak scores | MNI coordinates | Cluster size (voxels) |
| (P-values) | (x, y, z) | |||
| Bilateral Lingual Gyrus/Cerebellum Posterior Lobe | 17,18 | -5.12, (<0.01) | 0, -69, -3 | 498 |
Table 1: Significant differences in the ReHo values between the two groups. The GRF theory was used to set the statistical threshold at the voxel level for multiple comparisons (P < 0.01). ReHo = regional homogeneity; BA = Brodmann area; RP = retinitis pigmentosa; HC = health control; MNI = Montreal Neurological Institute; GRF = Gaussian random field.
| Condition | Brain regions | BA | Peak T scores | MNI coordinates | Cluster size (voxels) |
| (x, y, z) | |||||
| ROI in bilateral LGG/CPL | |||||
| RP > HC | Left thalamus | – | 3.1668 | -21, -18, -3 | 70 |
| RP > HC | Right thalamus | – | 3.5733 | 18, -24, 21 | 219 |
| RP < HC | Left Postcentral | – | -3.6226 | -48, -21, 39 | 262 |
Table 2: Comparison of seed-based FC values of the altered ReHo regions between the two groups. The GRF correction method was used to set the statistical threshold at the voxel level for multiple comparisons (P < 0.01). FC = functional connectivity; ReHo = regional homogeneity; ROI = region of interest; LGG = Lingual Gyrus; CPL = cerebellum posterior lobe; BA = Brodmann area; RP = retinitis pigmentosa; HC = health control; MNI = Montreal Neurological Institute; GRF = Gaussian random field.
Discussion
This report describes a protocol for computing ReHo and FC values for RP and HC groups and showed significantly different ReHo and FC values between the two groups. Notably, an important step in this process is the classification and screening of samples before the experiment. When we applied this protocol for our own analysis, all RP subjects were diagnosed by two experienced ophthalmologists. We excluded RP patients with other eye diseases such as glaucoma, cataracts, and optic atrophy. In addition, HCs enrolled in our study have no heart disease, cerebral diseases, or hypertension. The results show that ReHo values of the visual cortex and FC between the visual cortices and motor cortices decreased significantly in RP participants, as well as increased FC between the visual cortices and thalamus. Qin et al. demonstrated that RP patients showed a decreased FC density in V1, which is consistent with our findings.10 These findings may indicate the disconnection and reorganization of the intrinsic visual network of the retino-thalamocortical pathway and dorsal visual stream, suggesting visuospatial and stereoscopic vision impairment.
Another important issue in this protocol is the statistical analysis. When a two-sample t-test was performed to compare the ReHo and FC indexes using DPABI software, the effects of nuisance covariates (age, sex, and head motion parameters) were removed during statistical analysis.
There are some limitations in this ReHo method. In particular, neuropsychiatric scaling is not conducted in RP patients. Mental state and physiological noise might influence the accuracy of the results. Furthermore, the protocol does not compare brain structure differences between the RP and HC groups.
In addition to the applications described here, our combined ReHo and FC-based method provides promising potential approaches for assessment of intra- and interregional brain activity synchronizations. This method provides an efficient and practical means for capturing the precise inceptions of fMRI signals and generating reliable results using our data postprocessing approach. In addition, amplitude of low-frequency fluctuation11 and degree centrality12 enable measurement of regional activity changes in the resting state. In the future, multimodal MRI technologies will be used to determine the functional and morphological changes in RP patients. This technique may be useful in the clinical realm as a diagnostic tool for RP patients as further understanding is achieved regarding the complexities of the human neuronal system.
Declarações
The authors have nothing to disclose.
Acknowledgements
This research was supported by National Nature Science Foundation of China (NSFC, No. 81470628, 81800872); National Key R&D Program of China (No. 2017YFE0103400)
Materials
| BrainNet Viewer software | National Key Laboratory of Cognition Neuroscience and Learning, BNU | BrainNet Viwer 2013 | BrainNet Viewer is a brain network visualization tool to visualize structural and functional connectivity patterns |
| DPABI software | Institute of Psychology, CAS, Beijing, China | DPABI 4.3 | DPABI is a toolbox for data processing and analysis of brain imaging. |
| MATLAB | MathWorks, Natick, MA, USA | 2013a | MATLAB is a high-level technical computing language and interactive environment for algorithm development, data visualization, data analysis, and numeric computation. |
| MRI scanner | GE Healthcare, Milwaukee | MRI 3.0 | |
| SPM software | Wellcome Centre for Human Neuroimaging, UCL | SPM8 | SPM8 is a major update to the SPM software, containing substantial theoretical, algorithmic, structural and interface enhancements over previous versions. |
| SPSS | IBM, Chicago, IL, USA | SPSS version 20.0 | SPSS software platform offers advanced statistical analysis, text analysis, open-source extensibility, integration with big data and seamless deployment into applications. |
Referências
- Zang, Y., Jiang, T., Lu, Y., He, Y., Tian, L. Regional homogeneity approach to fMRI data analysis. NeuroImage. 22 (1), 394-400 (2004).
- Smith, R., et al. Resting state functional connectivity correlates of emotional awareness. NeuroImage. 159, 99-106 (2017).
- Hartong, D. T., Berson, E. L., Dryja, T. P. Retinitis pigmentosa. The Lancet. 368 (9549), 1795-1809 (2006).
- Ezquerra-Inchausti, M., et al. High prevalence of mutations affecting the splicing process in a Spanish cohort with autosomal dominant retinitis pigmentosa. Scientific Reports. 7 (1), 39652 (2017).
- Cunningham, S. I., Weiland, J. D., Bao, P., Lopez-Jaime, G. R., Tjan, B. S. Correlation of vision loss with tactile-evoked V1 responses in retinitis pigmentosa. Vision Research. 111, 197-207 (2015).
- Cunningham, S. I., Weiland, J. D., Pinglei, B., Tjan, B. S. . 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society. , 2841-2844 (2011).
- Ferreira, S., et al. Primary visual cortical remapping in patients with inherited peripheral retinal degeneration. NeuroImage: Clinical. 13, 428-438 (2017).
- Zhang, Y., et al. Reduced Field-of-View Diffusion Tensor Imaging of the Optic Nerve in Retinitis Pigmentosa at 3T. American Journal of Neuroradiology. 37 (8), 1510-1515 (2016).
- Yan, C. G., Wang, X. D., Zuo, X. N., Zang, Y. F. DPABI: Data Processing & Analysis for (Resting-State) Brain Imaging. Neuroinformatics. 14 (3), 339-351 (2016).
- Qin, W., Xuan, Y., Liu, Y., Jiang, T., Yu, C. Functional Connectivity Density in Congenitally and Late Blind Subjects. Cerebral Cortex. 25 (9), 2507-2516 (2015).
- Yu-Feng, Z., et al. Altered baseline brain activity in children with ADHD revealed by resting-state functional MRI. Brain and Development. 29 (2), 83-91 (2007).
- Zuo, X. N., et al. Network Centrality in the Human Functional Connectome. Cerebral Cortex. 22 (8), 1862-1875 (2012).

.
