Antibiotic Dereplication Using the Antibiotic Resistance Platform
Summary
We describe a platform that utilizes a library of isogenic antibiotic resistant Escherichia coli for the dereplication of antibiotics. The identity of an antibiotic produced by bacteria or fungi can be deduced by the growth of E. coli expressing its respective resistance gene. This platform is economically effective and time-efficient.
Abstract
One of the main challenges in the search for new antibiotics from natural product extracts is the re-discovery of common compounds. To address this challenge, dereplication, which is the process of identifying known compounds, is performed on samples of interest. Methods for dereplication such as analytical separation followed by mass spectrometry are time-consuming and resource-intensive. To improve the dereplication process, we have developed the antibiotic resistance platform (ARP). The ARP is a library of approximately 100 antibiotic resistance genes that have been individually cloned into Escherichia coli. This strain collection has many applications, including a cost-effective and facile method for antibiotic dereplication. The process involves the fermentation of antibiotic-producing microbes on the surface of rectangular Petri dishes containing solid medium, thereby allowing for the secretion and diffusion of secondary metabolites through the medium. After a 6 day fermentation period, the microbial biomass is removed, and a thin agar-overlay is added to the Petri dish to create a smooth surface and enable the growth of the E. coli indicator strains. Our collection of ARP strains is then pinned onto the surface of the antibiotic-containing Petri dish. The plate is next incubated overnight to allow for E. coli growth on the surface of the overlay. Only strains containing resistance to a specific antibiotic (or class) grow on this surface enabling rapid identification of the produced compound. This method has been successfully used for the identification of producers of known antibiotics and as a means to identify those producing novel compounds.
Introduction
Since the discovery of penicillin in 1928, natural products derived from environmental microorganisms have proven to be a rich source of antimicrobial compounds1. Approximately 80% of natural product antibiotics are derived from bacteria of the genus Streptomyces and other actinomycetes, while the remaining 20% is produced by fungal species1. Some of the most common antibiotic scaffolds used in the clinic such as the β-lactams, tetracyclines, rifamycins, and aminoglycosides, were originally isolated from microbes2. However, due to the rise of multidrug resistant (MDR) bacteria, our current panel of antibiotics has become less effective in treatment3,4. These include the “ESKAPE” pathogens (i.e., vancomycin-resistant enterococci and β-lactam-resistant Staphylococcus aureus, Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, and Enterobacter sp.), which are a subset of bacteria deemed to be associated with the highest risk by a number of major public health authorities such as the World Health Organization3,4,5. The emergence and global spread of these MDR pathogens results in a constant need for novel antibiotics3,4,5. Regrettably, the past two decades have demonstrated that the discovery of novel antibiotics from microbial sources is increasingly difficult6. Current approaches to drug discovery include the high-throughput screening of bioactive compounds, including natural product extract libraries, allowing for thousands of extracts to be tested at a given time2. However, once antimicrobial activity is detected, the next step is to analyze the contents of the crude extract to identify the active component and eliminate those containing known or redundant compounds7,8. This process, referred to as dereplication, is vital to prevent and/or significantly reduce the time spent on the re-discovery of known antibiotics7,9. Although a necessary step in natural product drug discovery, dereplication is notoriously laborious and resource-intensive10.
Ever since Beutler et al. first coined the term “dereplication”, extensive efforts have been made to develop innovative strategies for the rapid identification of known antibiotics11,12. Today the most common tools used for dereplication include analytical chromatographic systems such as high-performance liquid chromatography, mass spectrometry, and nuclear magnetic resonance-based detection methods11,13. Unfortunately, each of these methods requires the use of expensive analytical equipment and sophisticated data interpretation.
In an attempt to develop a dereplication method that can be rapidly performed without specialized equipment, we established the antibiotic resistance platform (ARP)10. The ARP can be used for the discovery of antibiotic adjuvants, the profiling of new antibiotic compounds against known resistance mechanisms, and the dereplication of known antibiotics in extracts derived from actinobacteria and other microbes. Here, we focus on its application in antibiotic dereplication. The ARP utilizes a library of isogenic Escherichia coli strains expressing individual resistance genes that are effective against the most commonly re-discovered antibiotics14,15. When the E. coli library is grown in the presence of a secondary metabolite-producing organism, the identity of the compound can be deduced by the growth of E. coli strains that express its associated resistance gene10. When the ARP was first reported, the library consisted of >40 genes conferring resistance to 16 antibiotic classes. The original dereplication template was designed to encompass a subset of resistance genes per antibiotic class to provide information regarding antibiotic subclass during the dereplication process. Today, the ARP is comprised of >90 genes that confer resistance to 18 antibiotic classes. Using our extensive collection of resistance genes, a secondary dereplication template has been developed and is known as the minimal antibiotic resistance platform (MARP). This template was created to eliminate gene redundancy and to simply provide information regarding the general antibiotic class that a dereplicated metabolite is related to. Additionally, the MARP template possesses both wildtype and a hyperpermeable/efflux deficient strain of E. coli BW25113 (E. coli BW25113 ΔbamBΔtolC), compared to the original incarnation of the ARP, which only utilizes the hyperpermeable strain. This unique aspect creates additional phenotypes during dereplication, indicating a compounds ability to cross the outer membrane of Gram-negative bacteria. Here, we describe a robust protocol to be followed when dereplicating with either the ARP and/or MARP, highlight the most critical steps to be followed, and discuss the various possible outcomes.
Protocol
1. Preparation of E. coli Library Glycerol Stocks (from Agar Slants)
- Streak the ARP/MARP E. coli strains from lysogeny broth (LB) agar slants onto Petri dishes containing LB agar and the appropriate selectable marker (Table 1).
- Prepare cultures for each of the E. coli strains by inoculating 3 mL of LB containing the appropriate selectable marker with a single colony. Grow overnight at 37 °C with aeration (250 rpm).
- Combine 800 μL of culture and 200 μL of sterile 80% glycerol in a 1.8 mL cryovial. Mix by inverting the tubes 3−4 times, and store at -80 °C.
2. ARP/MARP Frozen Stock Library Plate Preparation
- Streak the ARP/MARP strains from the glycerol stocks prepared in section 1 onto a new set of Petri dishes containing LB agar and the appropriate selectable marker. Grow overnight at 37 °C.
- Using aseptic technique, pipette 500 μL of cation adjusted Mueller Hinton broth (MHB) from a sterile reservoir into each well of a sterile 96 deep well plate.
- With the plates prepared in step 2.1, use an applicator stick to inoculate the 96 deep well plate in accordance with the ARP/MARP map (Supplemental Figure 1 and Supplemental Figure 2). Ensure that the appropriate selectable marker is added to each well. Place a breathable sealing membrane over the surface of the deep well plate and incubate overnight at 37 °C (250 rpm).
- Ensure that there are no contaminated wells by referring to the ARP/MARP map. Repeat if contaminated. Using a multi-channel pipettor, transfer 100 μL from each well of the deep well plate to a sterile 96-well round bottom plate. Repeat this step to create multiple frozen stock library plates.
NOTE: It is best to prepare at least 5 library plates at a time to keep from repeating steps 2.1−2.4 in the event of frozen stock library plate contamination. - Finish making the ARP/MARP frozen stock library plates by pipetting 100 μL of sterile 50% glycerol into each well and mix by gently pipetting up and down.
- Cover the plates with sterile aluminum seals and ensure that each well is individually sealed.
- Number the plates and dedicate only one frozen stock library plate for inoculating new templates at a given time. Keep the remainder as back-ups in the event of frozen stock library plate contamination.
- Place the plate lid on top of the aluminum seal and store at -80 °C.
3. Seed Culture and Dereplication Plate Preparation
- Using an applicator stick, inoculate 3 mL of Streptomyces antibiotic medium (SAM) (or other appropriate medium for the organism being tested) in a test tube containing one sterile glass bead (to break-up the mycelium) with the producing strain that is to be dereplicated. For Streptomyces, gently scrape spores from the surface of a colony.
- Using the same wooden applicator stick, streak a sterility control on a Petri dish containing Bennett’s agar.
- Incubate the seed culture at 30 °C with aeration for 6 days (250 rpm) and incubate the sterility control plate at 30 °C for 6 days.
NOTE: Refer to Table 2 for SAM and Bennett’s media recipes. The above instructions are suitable for most actinomycetes. Alter growth media as necessary for other bacteria and fungi. - Prepare dereplication plates by aspirating 23 mL of warm Bennett’s agar into a serological pipette and dispense 20 mL evenly across the surface of a rectangular Petri dish (Table of Materials), leaving the remainder of the medium in the pipette to prevent air bubble formation.
NOTE: Ensure that the surface being used to pour plates is level and perform this step before the agar has cooled too much; a flat surface is imperative for library pinning in the next stages. - Gently rotate the plate until the medium covers all areas of the plate and do not disturb it until the agar has set completely.
- Prepare nitrocellulose membrane sheets (Table of Materials) by using a rectangular Petri dish lid as a tracing template so that the sheets fit the surface of the dereplication plate. Cut the sheets and autoclave them in a sterile pouch.
NOTE: This membrane allows for organisms to sporulate on its surface, while secondary metabolites may be excreted into the medium below. Once grown, the membrane is removed to provide a clean surface for dereplication. The closer fit that the membrane paper has on the surface of the Bennett’s agar, the cleaner the dereplication result. - Check the sterility control plate to ensure that no contaminants are present after 6 days of incubation. If contamination-free, remove the lid of the rectangular Petri dish and pipette 200 μL of seed culture onto the surface of the Bennett’s agar.
- Evenly spread the culture across the surface of the entire plate using a sterile cotton swab.
- Place the nitrocellulose membrane prepared in step 3.6 over top of the culture on the surface of the Petri dish. Begin by aligning the bottom edge of the membrane to the bottom edge of the Petri dish, and slowly apply the membrane from the bottom edge to the top edge of the plate.
- Use a sterile cotton swab to smooth out any air bubbles that may have formed between the membrane-agar interface, ensuring that the membrane is flush to the agar.
- Put the lid back on the rectangular Petri dish and place it upside down in a sealed plastic bag. Incubate at 30 °C for 6 days.
4. Dereplication Plate MHB Overlay and ARP/MARP Library Plate Preparation
- After 6 days, remove the dereplication plate from the 30 °C incubator. Using sterile tweezers (autoclaved or sprayed thoroughly with 70% ethanol), carefully remove the nitrocellulose membrane from the surface of the Bennett’s agar.
NOTE: This step will remove the hydrophobic spores and mycelia grown on the surface of the membrane to provide a clean surface for dereplication, facilitating step 4.2. - As described for step 3.4, ensure the work surface is level and use a serological pipette to aspirate 23 mL of warm cation adjusted MHB agar. Create an overlay by dispensing 20 mL evenly across the surface of the dereplication plate, leaving the remainder of the medium in the pipette to prevent air bubble formation.
- Gently rotate the plate until the medium covers all areas and do not disturb it until the agar has set completely. Once cooled and solidified, return the dereplication plate to the sealed plastic bag and store it upside at 4 °C overnight.
NOTE: This step allows for diffusion of secondary metabolites from the fermented Bennett’s medium into the MHB agar overlay. If the nitrocellulose membrane was not prepared properly there will be spore growth around the edges of the plate, which have hydrophobic properties that repel the MHB agar. Do not pour the overlay on top of these spores because it can result in contamination of the overlay. - On the same day that the overlay is poured, inoculate a fresh ARP/MARP template by pipetting 100 μL of cation adjusted MHB into each well of a 96-well plate.
NOTE: To reduce the chance of spreading contamination during dereplication, use a single ARP/MARP library plate to only dereplicate 2−3 dereplication plates. Therefore, inoculate enough 96-well ARP/MARP plates based on the number of strains that will be dereplicated. - Take the frozen stock ARP/MARP library plate out of the -80 °C freezer. Remove the aluminum seal before condensation begins to form on its underside, thereby decreasing the chance of contaminating neighboring wells in the library plate.
- Using sterile 96-well pinning tools (or other forms of inoculation equipment), carefully pin from the frozen stock ARP/MARP library plate and inoculate the fresh MHB-containing 96-well plates. To minimize contamination during dereplication, prepare as many ARP or MARP library plates needed to only dereplicate 2−3 dereplication plates per library plate. Sterilize pinning tools between inoculating each plate.
- Put a new sterile aluminum seal on the frozen template once complete and return it to the -80 °C freezer. Place the inoculated 96-well plates inside of a sealed plastic bag and incubate at 37 °C with aeration (250 rpm) for 18 h.
NOTE: New frozen stock library plates can be prepared from this step after ensuring that no contamination is present. Add glycerol to the plate before storing at -80 °C as described in step 2.5.
5. Dereplicating Using the ARP/MARP
- Remove the ARP/MARP template from the incubator and ensure that no contaminants are present. Always dereplicate using a template that is freshly prepared and not directly from the frozen stock.
- Remove the dereplication plates from 4 °C and allow to equilibrate to room temperature. If there is condensation, open the lids and allow to dry in a sterile environment.
- Using sterile pinning tools (or other inoculation equipment), pin from the ARP/MARP library plate onto the surface of the MHB agar overlay of the dereplication plates. Be careful not to pierce the agar. Sterilize pinning tools in between inoculating each dereplication plate.
- After pinning the template onto the surface of the dereplication plates, allow the template inoculum to dry for 3−5 min. Place the inoculated dereplication plates upside down in a sealed plastic bag and incubate overnight at 37 °C.
- Analyze dereplication results the following day by comparing growth on the dereplication plate to wells that correspond to the ARP/MARP map (Table 3 and Table 4).
Representative Results
The following results were obtained when a collection of antibiotic-producing strains of interest were dereplicated using the ARP and/or MARP.
A diagram of the ARP/MARP dereplication workflow is depicted in Figure 1, and library plate maps are shown in Supplemental Figure 1 and Supplemental Figure 2. Figure 2 demonstrates a positive dereplication result wherein the environmental extract WAC 8921 is identified as a chloramphenicol producer. Figure 3 shows a lack of ARP growth entirely, which indicates the presence of either an unknown antibiotic or a less commonly found antibiotic that is not accounted for in the ARP/MARP library plate. Figure 4 demonstrates a growth pattern that is unique to the MARP because of its utilization of both wildtype E. coli BW25113 and a hyperpermeable and efflux deficient mutant E. coli BW25113 ΔbamBΔtolC. This result suggests the presence of a compound with antimicrobial activity that is unable to surpass an intact outer membrane. Figure 5 shows an E. coli growth pattern that suggests the improper sterilization of pinning tools and Figure 6 shows an example of ARP/MARP frozen stock library plate contamination. Figure 7 demonstrates what happens if the agar overlay is pierced during dereplication. Lastly, Figure 8 shows MHB overlay related contamination that can occur during the dereplication process.
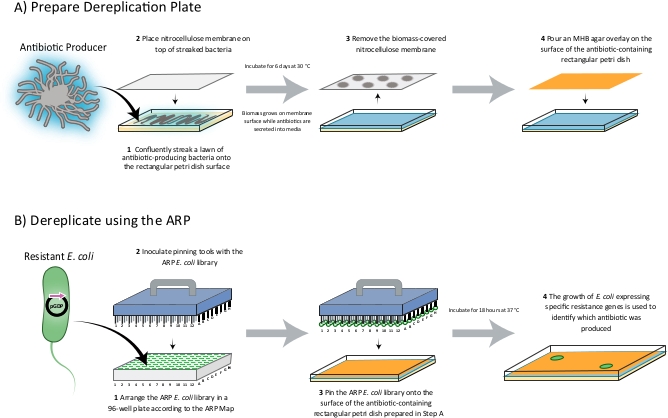
Figure 1: A schematic of the dereplication process. The producing strain to be dereplicated is streaked onto a rectangular Petri dish as a lawn and a nitrocellulose membrane is placed on top. The plate is then incubated for 6 days wherein the producing-strain related biomass grows on the surface of the membrane while and secondary metabolites produced are secreted into the Petri dish media. After a 6-day fermentation period, the membrane is removed and an MHB overlay is added to the surface of the antibiotic-containing media to provide a smooth surface for pinning. The ARP/MARP E. coli library, which is arranged in a 96-well plate format according to the ARP/MARP Maps, is then pinned onto the surface of the overlay. After incubating the tray overnight at 37 °C, the growth of E. coli strains expressing specific resistance genes indicates the identity of the compound produced. Please click here to view a larger version of this figure.

Figure 2: Dereplication of a known antibiotic. The producing strain WAC 8921 was dereplicated using the ARP template. Growth of E. coli BW25113 ΔbamBΔtolC pGDP1:CAT on the surface of the MHB agar overlay indicates that WAC 8921 is a chloramphenicol producer. Please click here to view a larger version of this figure.
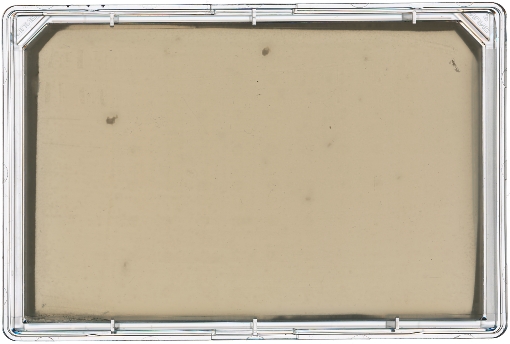
Figure 3: Dereplication of an unknown antibiotic. The producing strain WAC 9941 was dereplicated using the ARP template. A lack of E. coli library growth was seen on the surface of the rectangular Petri dish, indicating that either WAC 9441 is producing an unknown antimicrobial compound or a rare antibiotic that is not accounted for in the ARP. Please click here to view a larger version of this figure.

Figure 4: Identification of an antimicrobial compound that cannot traverse an intact outer membrane. The producing strain WAC 4178 was dereplicated using the MARP template. Strains of E. coli BW25113 are capable of growing on the surface of the secondary metabolite-containing media, whereas all strains of E. coli BW25113 ΔbamBΔtolC cannot grow. This suggests that WAC 4178 is producing an antimicrobial compound that cannot traverse an intact outer membrane. Please click here to view a larger version of this figure.
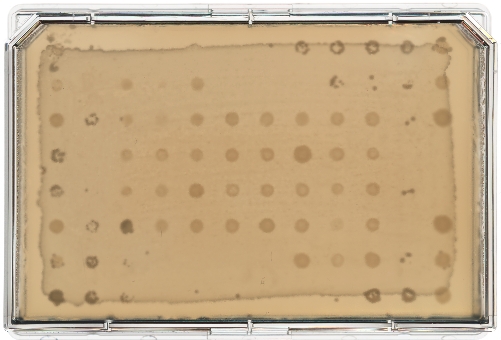
Figure 5: Contamination due to non-sterile pinning tools. The producing strain WAC 7094 was dereplicated using the ARP template. The presence of E. coli library growth in areas that did not have an assigned E. coli strain suggests that the pinning tools used to inoculate the MHB agar overlay of the rectangular Petri dish were not properly sterilized. This results in the transfer of unknown E. coli strains across the overlay. Please click here to view a larger version of this figure.
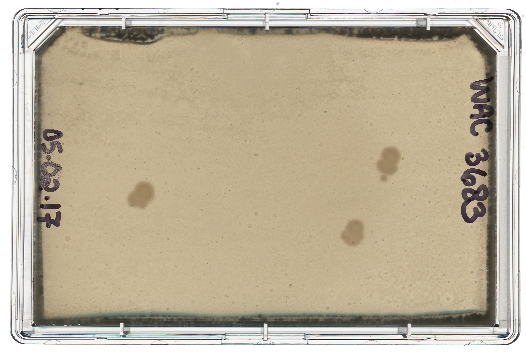
Figure 6: Contamination due to a contaminated frozen stock ARP/MARP template. The producing strain WAC 3683 was dereplicated using the ARP template. Three distinct E. coli colonies grew on the rectangular Petri dish surface: two correspond with E. coli BW25113 ΔbamBΔtolC expressing STAT, a streptothricin resistance enzyme, and the other corresponds to E. coli BW25113 ΔbamBΔtolC expressing VIM-2ss, a β-lactam resistance enzyme. Due to a lack of replicating blaVIM2ss colony growth, in addition to the lack of cross-resistance known to occur between these two antibiotic classes, it can be assumed that a strain other than E. coli ΔbamBΔtolC pGDP1: blaVIM2ss is growing in the respective frozen library plate well. Please click here to view a larger version of this figure.
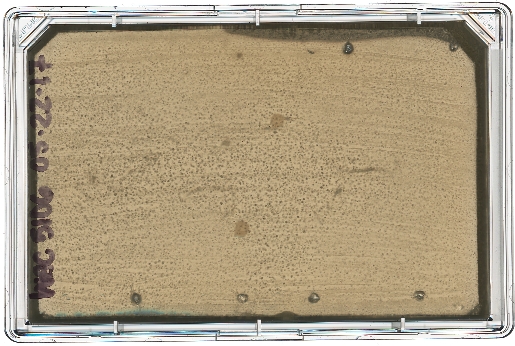
Figure 7: Pierced MHB agar overlay. The producing strain WAC 5106 was dereplicated using the ARP template. This strain was found to be a streptomycin producer, as indicated by the growth of E. coli BW25113 ΔbamBΔtolC pGDP3:aph(6)-Ia. Puncture holes can be seen on the surface of the MHB agar overlay along the perimeter of the plate. While this does not affect the dereplication results, it can make the data difficult to interpret at first glance. Please click here to view a larger version of this figure.

Figure 8: Contamination of the MHB agar overlay. Contaminated MHB agar produces an irregular growth pattern on the surface of the overlay that becomes visible after incubating the plate overnight at 37 °C. Although E. coli growth may still be visible through the contamination, it is advised to repeat the experiment before extrapolating data from the plate. Please click here to view a larger version of this figure.
| Plasmid | Selectable Marker |
| pGDP1 | Kanamycin 50 μg/mL |
| pGDP2 | |
| pGDP3 | Ampicillin 100 μg/mL |
| pGDP4 | |
| None | – |
Table 1: Selectable markers used in the pGDP plasmid series. Streak the ARP/MARP E. coli strains onto LB agar Petri dishes containing the appropriate selectable marker at the right concentration for each plasmid.
| Media | Ingredient | Amount |
| SAM | Glucose | 15 g |
| Soya peptone | 15 g | |
| NaCl | 5 g | |
| Yeast extract | 1 g | |
| CaCO3 | 1 g | |
| Glycerol | 2.5 mL | |
| ddH2O | To 1 L | |
| Bennett's | Potato starch | 10 g |
| Casamino acids | 2 g | |
| Yeast extract | 1.8 g | |
| Czapek mineral mix | 2 mL | |
| Agar (optional) | 15 g | |
| ddH2O | To 1 L | |
| Czapek mineral mix | KCl | 10 g |
| MgSO4∙7H2O | 10 g | |
| NaNO3 | 12 g | |
| FeSO4∙7H2O | 0.2 g | |
| Concentrated HCl | 200 µL | |
| ddH2O | To 100 mL |
Table 2: Recipes for SAM and Bennett’s media, and Czapek mineral mix. Adjust SAM and Bennett’s to pH 6.8 before autoclaving, and filter sterilize the Czapek mineral mix.
| Antibiotic Class | Antibiotic | Resistance Gene | E. coli Strain | Well Position |
| Aminoglycosides | Streptomycin | aph(3’’)-Ia | ΔbamBΔtolC BW25113 | B3, G10 |
| 2- Deoxystreptamine | rmtB | ΔbamBΔtolC BW25113 | F3, C10 | |
| Apramycin | apmA | ΔbamBΔtolC BW25113 | C5, F8 | |
| Spectinomycin | aph(9)-Ia | ΔbamBΔtolC BW25113 | B5, G8 | |
| β-lactams | Penicillin | NDM-1 | ΔbamBΔtolC BW25113 | B4, G9 |
| Cephalosporin | ||||
| Carbapenam | ||||
| Lincosamides | Lincosamides | ermC | ΔbamBΔtolC BW25113 | D4, E9 |
| Macrolides | Macrolides | ermC | ||
| Type B Streptogramins | Type B Streptogramins | ermC | ||
| Type A Streptogramins | Type A Streptogramins | vatD | ΔbamBΔtolC BW25113 | C3, F10 |
| Streptothricin | Streptothricin | STAT | ΔbamBΔtolC BW25113 | D3, E10 |
| Tetracyclines | Tetracycline | tet(A) | ΔbamBΔtolC BW25113 | D5, E8 |
| Chloramphenicols | Chloramphenicols | CAT | ΔbamBΔtolC BW25113 | E4, D9 |
| Fosfomycins | Fosfomycins | fosA | ΔbamBΔtolC BW25113 | F6, C7 |
| Rifamycins | Rifamycins | arr | ΔbamBΔtolC BW25113 | E3, D10 |
| Polymyxins | Polymyxins | MCR-1 | wild-type BW25113 | C6, F7 |
| Echinomycins | Echinomycins | uvrA | ΔbamBΔtolC BW25113 | F4, C9 |
| Sideromycins | Albomycin | fhuB mutant | ΔbamBΔtolC BW25113 | C4, F9 |
| Tuberactinomycins | Viomycin | vph | ΔbamBΔtolC BW25113 | F5, C8 |
| N/A | N/A | N/A | wild-type BW25113 | C1, C12, F1, F12, E5, D8 |
| N/A | N/A | N/A | ΔbamBΔtolC BW25113 | A1, A12, B1, B12, D6, D7, E6, E7, G1, G12, H1, H12 |
Table 3: Well designation table for the minimal ARP strains. This table indicates which well of a 96-well plate that each of the minimal ARP strains can be found in according to the minimal ARP library plate map. The table also lists which antibiotic class each gene confers resistance to. Please note that some genes may confer resistance to more than one antibiotic within the given antibiotic class.
| Antibiotic Class | Antibiotic | Resistance Gene | E. coli Strain | Well Position |
| Aminoglycosides | Streptomycin | aph(3’’)-Ia | ΔbamBΔtolC BW25113 | B2, G11 |
| aph(6)-Ia | ΔbamBΔtolC BW25113 | C6,F7 | ||
| Spectinomycin | aph(9)-Ia | ΔbamBΔtolC BW25113 | A2, H11 | |
| Gentamicin | aac(3)-Ia | ΔbamBΔtolC BW25113 | A3,H10 | |
| ant(2’’)-Ia | ΔbamBΔtolC BW25113 | A5, H8 | ||
| aph(2’’)-Id | ΔbamBΔtolC BW25113 | A4, H9 | ||
| armA | ΔbamBΔtolC BW25113 | A6, H7 | ||
| aac(6’)-aph(2’’)-Ia | ΔbamBΔtolC BW25113 | B5,G8 | ||
| Kanamycin | aph(3’)-Ia | ΔbamBΔtolC BW25113 | B4, G9 | |
| aph(3’)-Illa | ΔbamBΔtolC BW25113 | B3, G10 | ||
| Hygromycin | aph(4)-Ia | ΔbamBΔtolC BW25113 | B6,G7 | |
| β-lactams | Amoxicillin | TEM-1 | ΔbamBΔtolC BW25113 | F6, C7 |
| Ceftazidime | CTX-M-15 | ΔbamBΔtolC BW25113 | F5, C8 | |
| Oxacillin | OXA-10 | ΔbamBΔtolC BW25113 | G5, B8 | |
| OXA-48 | ΔbamBΔtolC BW25113 | H5, A8 | ||
| Meropenem | IMP-7ss | ΔbamBΔtolC BW25113 | G4, B9 | |
| KPC-2 | ΔbamBΔtolC BW25113 | G6, B7 | ||
| NDM-1 | ΔbamBΔtolC BW25113 | H6, A7 | ||
| Imipenem | VIM-2 | ΔbamBΔtolC BW25113 | F4, C9 | |
| Lincosamides | Lincosamides | ermC | ΔbamBΔtolC BW25113 | C4, F9 |
| lnu(A) | ΔbamBΔtolC BW25113 | C5, F8 | ||
| Macrolides | Macrolides | ermC | ΔbamBΔtolC BW25113 | C4, F9 |
| mphA | ΔbamBΔtolC BW25113 | C3, F10 | ||
| mphB | ΔbamBΔtolC BW25113 | C2, F11 | ||
| Type B Streptogramins | Type B Streptogramins | ermC | ΔbamBΔtolC BW25113 | C4, F9 |
| Vgb | ΔbamBΔtolC BW25113 | D4, E9 | ||
| Type A Streptogramins | Type A Streptogramins | vatD | ΔbamBΔtolC BW25113 | D5, E8 |
| Streptothricin | Streptothricin | STAT | ΔbamBΔtolC BW25113 | D3, E10 |
| Tetracyclines | Tetracycline | tet(M) | ΔbamBΔtolC BW25113 | E5, D8 |
| Chloramphenicols | Chloramphenicols | CAT | ΔbamBΔtolC BW25113 | E4, D9 |
| Fosfomycins | Fosfomycins | fosA | ΔbamBΔtolC BW25113 | H4, A9 |
| Rifamycins | Rifamycins | arr | ΔbamBΔtolC BW25113 | E3, D10 |
| N/A | N/A | N/A | ΔbamBΔtolC BW25113 | A1, A12, B1, B12, D6, D7, E6, E7, G1, G12, H1, H12 |
Table 4: Well designation table for the ARP strains. This table indicates which well of a 96-well plate that each of the ARP strains can be found in according to the ARP library plate map. The table also lists which antibiotic class each gene confers resistance to. Please note that some genes may confer resistance to more than one antibiotic within the given antibiotic class.
Supplemental Figure 1: Library plate map used for the original antibiotic resistance platform (ARP) template. Organize the respective E. coli strains in a 96-well plate using this format to ensure that all necessary controls and duplicates are included. This figure has been modified from Cox et al.10. Please click here to download this figure.
Supplemental Figure 2: Library plate map used for the minimal antibiotic resistance platform (MARP) template. Organize the respective E. coli strains in a 96-well plate using this format to ensure that all necessary controls and duplicates are included. Please click here to download this figure.
Discussion
The protocol described above can be applied to both the discovery of novel antimicrobial compounds and adjuvants that can be used in conjunction with existing antibiotics to rescue their activity. The platform takes advantage of the high substrate specificity of resistance mechanisms and their cognate antibiotics, to dereplicate compounds within crude natural product extracts. Although the time required for dereplication plates to be prepared is lengthy (~2 weeks), the dereplication process itself is complete after a single overnight incubation period, which is rapid in comparison to the time it can take to isolate and characterize a compound from crude extracts. Additionally, no expensive or highly specialized equipment is required, making this platform accessible and cost-effective.
Another major benefit of this platform is its flexibility. The ARP can be expanded to contain resistance genes that encompass more antibiotic classes. This is achieved by monitoring literature for the emergence of novel resistance enzymes and using basic molecular cloning techniques to add the genes to the E. coli library. Furthermore, the dereplication template is customizable based on the desired level of broad or narrow range substrate specificity that an individual wants to use when dereplicating. Any combination of genes in the E. coli library can be used to make novel library plates with the ability to detect compounds with different profiles. For example, a β-lactamase expressing E. coli template could be developed to allow for the highly specific dereplication of β-lactams and their different subclasses.
While this platform was initially designed to dereplicate compounds on solid media, it also been works in liquid media. This is useful when working with compounds that have already been purified wherein only a limited amount is available for testing, or when working with compounds that do not diffuse easily or consistently in solid media. Lastly, while this protocol was described using the E. coli strains BW25113 and BW25113 ΔbamBΔtolC, the platform can be used with the resistance gene library expressed in different strains of E. coli (dereplication phenotypes may vary). Ultimately, the Antibiotic Resistance Platform is flexible, has many applications, and is advantageous over other dereplication methods.
To ensure that reproducible and non-contaminated results are obtained, it is critical to follow appropriate sterilization and aseptic techniques. Failure to do so will result in contamination of the pinning tools, library plate, or the dereplication plate itself. While selectable markers are present in the E. coli library, which can help prevent contamination caused by bacteria missing the marker, it does not prevent the cross-contamination of strains using the same marker. To reduce the risk of this happening it is essential to carefully sterilize the bacterial pinning tools before pinning from the library plate. Each library plate should only be pinned from 3−4 times maximum before discarding for a new template. This prevents the spread of contamination across all dereplication plates if a library plate becomes contaminated during the pinning process. Additionally, no antibiotics are used when inoculating the dereplication plate and so great care must be taken to prevent contamination before it is left to ferment for six days. Another possible source of contamination is in the MHB agar overlay of the dereplication plate. If the overlay media is contaminated, growth will only appear after the overnight incubation at 37 °C post-pinning. Overlay contamination can make it extremely difficult to analyze the growth of the E. coli library on the overlay surface. To reduce the chances of overlay contamination, prepare MHB agar fresh before pouring the overlay. It is recommended that until an individual is comfortable with this method, dereplication plates should always be prepared in duplicate or triplicate such that in the event of contamination of one plate, data can still be extracted from the hopefully non-contaminated plates.
Lastly, it is important to note that the ARP/MARP has limitations. This protocol is not suitable for the de-replication of producing-strains that produce more than one bioactive compound. Each strain in the E. coli library has been designed to express a single resistance gene. If two antibiotics are being produced by an organism, neither resistance gene will confer resistance to the second antibiotic, thereby resulting in cell death of both strains. Thus, this possibility must be considered when dereplication results suggest the presence of a novel antibiotic because the production of multiple antibiotics cannot be detected by the current single construct E. coli library. One approach that can be taken to combat the challenge of dereplicating strains that produce more than one antibiotic involves using the agar-plug procedure described in the original ARP paper by Cox et al.10. In this method, a portion of a fermented solid medium is removed from an antibiotic-producer containing plate and placed onto a lawn of indicator ARP strain. The indicator strain can be any of the resistant E. coli strains in the ARP library. Zones of inhibition are then used to compare the bioactivity of a producing-strain against the ARP strains and a wild-type strain. ARP strains that form an inhibitory zone of decreased size compared to the wild-type strain can resist the compound being produced. This method has proven to be effective at identifying strains capable of producing multiple antibiotics10.
In summary, for obtaining the best results when dereplicating with the ARP/MARP it is recommended that dereplication plates are prepared in duplicate or triplicate. Other critical steps in the protocol include pinning from a fresh library plate (never frozen) and removing as much biomass as possible from the producing-strain during the membrane removal stage. If all necessary steps are followed, one should have successful dereplication results for a producing-strain of interest within a two-week time frame.
Declarações
The authors have nothing to disclose.
Acknowledgements
Research in the Wright lab pertaining to the ARP/MARP was supported by the Ontario Research Fund and Canadian Institutes of Health Research grant (FRN-148463). We would like to acknowledge Sommer Chou for assisting in the expansion and organization of the ARP library.
Materials
| Agar | Bio Shop | AGR003.5 | |
| AlumaSeal CS Films for cold storage | Sigma-Aldrich | Z722642-50EA | |
| Ampicillin Sodium Salt | Bio Shop | AMP201.100 | |
| BBL Mueller Hinton II Broth (Cation-Adjusted) | Becton Dickinson | 212322 | |
| BBL Phytone Peptone (Soytone) | Becton Dickinson | 211906 | |
| Calcium Carbonate | Bio Shop | CAR303.500 | |
| Casamino acid | Bio Basic | 3060 | |
| Cotton-Tipped Applicators | Fisher Scientific | 23-400-101 | |
| CryoPure Tube 1.8ml mix.colour | Sarstedt | 72.379.992 | |
| D-glucose | Bio Shop | GLU501.5 | |
| Disposable Culture Tube, 16x100mm | Fisher Scientific | 14-961-29 | |
| Ethyl Alcohol Anhydrous | Commercial Alcohols | P016EAAN | |
| Glass Beads, Solid | Fisher Scientific | 11-312C | |
| Glycerol | Bio Shop | GLY001.4 | |
| Hydrochloric Acid | Fisher Scientific | A144-212 | |
| Instant sealing sterilization pouch | Fisher Scientific | 01-812-54 | |
| Iron (II) Sulfate Heptahydrate | Sigma-Aldrich | F7002-250G | |
| Kanamycin Sulfate | Bio Shop | KAN201.50 | |
| LB Broth Lennox | Bio Shop | LBL405.500 | |
| Magnesium Sulfate Heptahydrate | Fisher Scientific | M63-500 | |
| MF-Millipore Membrane Filter, 0.45 µm pore size | Millipore-Sigma | HAWP00010 | 10 FT roll, hydrophillic, white, plain |
| Microtest Plate 96 well, round base | Sarstedt | 82.1582.001 | |
| New Brunswick Innova 44 | Eppendorf | M1282-0000 | |
| Nunc OmniTray Single-Well Plate | Thermo Fisher Scientific | 264728 | with lid, sterile, non treated |
| Petri dish 92x16mm with cams | Sarstedt | 82.1473.001 | |
| Pinning tools | ETH Zurich | – | Custom order |
| Potassium Chloride | Fisher Scientific | P217-500 | |
| Potato starch | Bulk Barn | 279 | |
| Sodium Chloride | Fisher Scientific | BP358-10 | |
| Sodium Nitrate | Fisher Scientific | S343-500 | |
| Wood Applicators | Dukal Corporation | 9000 | |
| Yeast Extract | Fisher Scientific | BP1422-2 |
Referências
- Lo Grasso, L., Chillura Martino, D., Alduina, R., Dhanasekaran, D., Jiang, Y. Production of Antibacterial Compounds from Actinomycetes. actinobacteria. Basics and Biotechnological Applications. , (2016).
- Thaker, M. N., et al. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nature Biotechnology. 31, 922-927 (2013).
- Gajdács, M. The Concept of an Ideal Antibiotic: Implications for Drug Design. Molecules. 24, 892 (2019).
- Boucher, H. W., et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clinical Infectious Diseases. 48, 1-12 (2009).
- Gajdács, M. The Continuing Threat of Methicillin-Resistant Staphylococcus aureus. Antibiotics. 8, 52 (2019).
- Gaudêncio, S. P., Pereira, F. Dereplication: Racing to speed up the natural products discovery process. Natural Product Reports. 32, 779-810 (2015).
- Ito, T., Masubuchi, M. Dereplication of microbial extracts and related analytical technologies. The Journal of Antibiotics (Tokyo). 67, 353-360 (2014).
- Van Middlesworth, F., Cannell, R. J. Dereplication and Partial Identification of Natural Products. Methods in Biotechnology. , 279-327 (2008).
- Tawfike, A. F., Viegelmann, C., Edrada-Ebel, R., Roessner, U., Dias, D. A. Metabolomics and Dereplication Strategies in Natural Products. Metabolomics Tools for Natural Product Discovery: Methods and Protocols. , 227-244 (2013).
- Cox, G., et al. A Common Platform for Antibiotic Dereplication and Adjuvant Discovery. Cell Chemical Biology. 24, 98-109 (2017).
- Hubert, J., Nuzillard, J. M., Renault, J. H. Dereplication strategies in natural product research: How many tools and methodologies behind the same concept. Phytochemistry Reviews. 16, 55-95 (2017).
- Beutler, J. Dereplication of phorbol bioactives: Lyngbya majuscula and Croton cuneatus. Journal of Natural Products. 53, 867-874 (1990).
- Mohimani, H., et al. Dereplication of microbial metabolites through database search of mass spectra. Nature Communications. 9, 1-12 (2018).
- Baltz, R. H. Marcel Faber Roundtable: Is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration. Journal of Industrial Microbiology and Biotechnology. 33, 507-513 (2006).
- Baltz, R. H. Antibiotic discovery from actinomycetes: Will a renaissance follow the decline and fall. Archives of Microbiology. 55, 186-196 (2005).

