NF-κB-dependent Luciferase Activation and Quantification of Gene Expression in Salmonella Infected Tissue Culture Cells
Summary
Here, we present a protocol to quickly and easily measure nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) activation in cell lines expressing NF-κB::luciferase reporter constructs, via measurements of luminescence in the cell lysate. Additionally, gene expression is determined via RT-qPCR isolated from cells infected with Salmonella Typhimurium.
Abstract
The dimeric transcription factor NF-κB regulates many cellular response pathways, including inflammatory pathways by inducing the expression of various cytokines and chemokines. NF-κB is constitutively expressed and is sequestered in the cytosol by the inhibitory protein nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha (IκBα). Activation of NF-κB requires the degradation of IκBα, which then exposes a nuclear localization signal on NF-κB and promotes its trafficking to the nucleus. Once in the nucleus, NF-κB binds to the promotor region of NF-κB target genes such as interleukin 6 (IL-6) and IL-23, to promote their expression.
The activation of NF-κB occurs independently of transcription or translation. Therefore, the activation state of NF-κB must be measured either by quantifying NF-κB specifically in the nucleus, or by quantifying expression of NF-κB target genes. In this protocol, cells stably transfected with an NF-κB::luciferase reporter construct are assayed for NF-κB activation using in vitro tissue culture techniques. These cells are infected with Salmonella Typhimurium to activate NF-κB, which traffics to the nucleus and binds to κB sites in the promoter region of luciferase, inducing its expression. Cells are lysed and analyzed with the luciferase assay system. The amount of luciferase produced by the cells correlates with the intensity of the luminescence signal, which is detected by a plate reader. The luminescence signal generated by this procedure provides a quick and highly sensitive method by which to assess NF-κB activation under a range of conditions. This protocol also utilizes quantitative reverse transcription PCR (RT-qPCR) to detect relative mRNA levels that are indicative of gene expression.
Introduction
The nuclear factor-κB (NF-κB) family of proteins are important transcription activators that regulate gene expression in various biological pathways. Activation of NF-κB induces transcription of target genes, many of which are important for immune and inflammatory responses, cell proliferation, stress responses and cancer progression1,2. NF-κB plays an integral role in mediating early inflammatory outcomes for pathogen clearance. Given the many biological processes mediated by NF-κB activation, disruptions in its signaling can have serious consequences for health and disease. Loss of function mutations in NF-κB signaling are associated in several immune deficiency phenotypes, while gain of function mutations are associated with several types of cancers, including B-cell lymphomas and breast cancers3. Additionally, many pathogens have been shown to directly modulate the activation state of NF-κB through expression of virulence factors4,5,6,7.
Activation of NF-κB is known to be a consequence of many variable stimuli including bacterial products such as lipopolysaccharides (LPS), flagellin and peptidoglycans known as pathogen-associated molecular patterns (PAMPs). These PAMPs are detected by pattern recognition receptors (PRRs) such as the Toll-like receptors (TLRs) and Nod-like receptors (NLRs) leading to the activation of NF-κB and the subsequent expression of an array of NF-κB-dependent inflammatory genes8. In addition to PRR activation by PAMPs, other bacterial products, such as bacterial effector proteins, can induce the activation of NF-κB. Interestingly, bacteria also express effector proteins that actively attenuate the NF-κB pathway and enhance their pathogenicity, underscoring the importance of NF-κB as an essential mediator of immunity9.
There are five different subunits that form the NF-κB dimers; p50, p52, RelA (p65), RelB and cRel. The two main NF-κB heterodimers are the p50:RelA and the p52:RelB dimers. The activated NF-κB dimers bind to DNA sites, known as κB sites, in the promoter and enhancer regions of various target genes. Under normal homeostatic conditions, NF-κB interacts with a family of inhibitor proteins known as IκB proteins to remain inactive. Upon stimulation, IκB is phosphorylated by IκB Kinase (IKK), which allows it to be targeted for ubiquitination, and subsequently degradation. Degradation of IκB activates NF-κB by revealing a nuclear localization signal. NF-κB then translocates to the nucleus, where it binds κB sites in the promoter region of target genes and promote transcription10. Thus, activation of NF-κB upregulates mRNA expression of NF-κB target genes, and this change can be measured through RNA quantification assays such as RT-qPCR11.
Several methods exist and are commonly used for the measurement of NF-κB activation, including electrophoretic mobility shift assays (EMSA), nuclear translocation, and gene reporter assays. EMSA is used to detect protein complexes with nucleic acids. Stimulated cells are fractionated to isolate nuclear proteins, including the translocated NF-κB, which is then incubated with radiolabeled nucleotides containing the NF-κB binding domain. The samples are run on a gel and imaged by autoradiography of 32P-labeled nucleic acid. If NF-κB is present in the protein fraction, it will bind the nucleotides, which will migrate slower through the gel and present as discrete bands. Nuclear fractions of cells lacking activated NF-κB (e.g., unstimulated control cells) will produce no bands as the nucleotides migrate faster to the end of the gel. A major drawback of this method is that it is largely quantitative in the binary sense (i.e., on or off) and does not adequately capture meaningful differences in NF-κB binding capacity. Additionally, this method does not consider chromatin structures that are functionally important for NF-κB target genes12,13.
Similar to the previous method, there is a "non-shift" assay in which multi-well plates are coated with nucleotides containing the NF-κB binding sequence. Following treatment of cells with nuclear fractions of protein, NF-κB will bind to the nucleotides bound to the well. Anti-NF-κB antibodies are then added, which will interact with the bound NF-κB and produce a colorimetric signal proportional to the amount of NF-κB, indicating the degree of NF-κB activation. This method is advantageous over the EMSA in that it does not require radiolabeled nucleic acids and is quantitative, in comparison. However, a caveat of this method is that it again does not differentiate between chromatin states of NF-κB target genes14.
Another method by which NF-κB activation may be detected is by chromatin immunoprecipitation (ChIP), whereby DNA and interacting proteins are cross-linked with formaldehyde and immunoprecipitated with specific anti-NF-κB antibodies. The specific nucleotide fragments are then purified and identified through PCR amplification or direct high throughput sequencing. Results generated from this method provide semi-quantitative results of NF-κB binding activity with target genes. However, the results are highly dependent on the fixation conditions and purification processes at each step15.
In nuclear translocation assays, cells are stimulated to induce NF-κB activation and then fixed. Anti-p65 antibodies are added to fixed cells. Alternatively, the p65 subunit itself can be tagged with a fluorescent peptide such as green fluorescent green (GFP). In either case, immunofluorescence will allow imaging of localization of p65 to determine cellular distribution. By measuring the proportion of cytosolic and nuclear localized protein, investigators can determine the relative activation state of NF-κB. A drawback of this method is that the immunofluorescence is comparatively time consuming, requires expensive antibodies, and needs relatively greater technical expertise16.
Reporter genes are commonly used tools to study the regulatory and expression patterns of a gene of interest. Typically, reporter genes are constructed from the promoter sequence of a gene of interest fused to a gene coding for an easily detectable protein. Proteins with enzymatic activities, fluorescence, or luminescence properties are commonly chosen for their ability to be assayed and quantified. Thus, the read-out (e.g., luminescence, fluorescence) serves as a signal for detection of the gene expression. These reporter constructs can then be introduced into different cell types, such as epithelial cells or macrophages.
Described in the protocol is the use of a cloned HeLa cell line (HeLa 57A) that is stably transfected with a luciferase reporter containing three copies of the κB consensus of the immunoglobulin κ-chain promoter region17. Expression of luciferase is dependent on the activation of NF-κB, which occurs following cell stimulation. Stimulated cells are easily lysed using cell lysis buffer provided in the luciferase assay kit. A portion of the cell lysate is then mixed with luciferase assay buffer that contains luciferin. Luciferin is the substrate of luciferase and is required for the generation of light in the presence of luciferase. After combining the assay buffer with the lysate, the solution will emit light in a process known as luminescence. The amount of light produced, given in lumens, is proportional to the amount of luciferase present in the lysate and serves as a measure of NF-κB activation. The lumen readings are interpreted in comparison to an unstimulated standard to account for baseline NF-κB activity and the signal itself is stable for several minutes to allow for reliable measurement. In addition, the HeLa 57A cell line is stably transfected with a NF-κB-independent β-galactosidase reporter. The β-galactosidase reporter is constitutively expressed, and β-galactosidase activity can be measured to control for cell viability or variation in cell numbers17. The luciferase values can then be adjusted to the β-galactosidase values and reported as fold increase over the unstimulated control cells.
Since NF-κB is a transcription factor responsible for the increased expression of NF-κB-dependent target genes, a follow up experiment to control for NF-κB-dependent increased gene expression is quantitative reverse transcription polymerase chain reaction (RT-qPCR). RT-qPCR is a highly sensitive method by which changes in the gene expression can be quantified over several orders of magnitude. Stimulated and control cells are harvested for RNA via phenol-chloroform extraction. Following phase separation, RNA is extracted as the major component of the aqueous layer. RNA is then precipitated and washed to produce a pure pellet. This pellet is then reconstituted and further cleaned of contaminant DNA via DNase treatment. The pure RNA is then reverse transcribed to create complementary DNA (cDNA). This cDNA can then be analyzed through quantitative PCR techniques, where the abundance of a specific mRNA sequence is quantified to determine gene expression. This technique does not elucidate translational control, post translational modification, protein abundance, or protein activity. However, many genes, particularly those involved in pro-inflammatory processes, are regulated via NF-κB and their mRNA abundance is indicative of their expression.
The method proposed here utilizes a fast and simple way by which NF-κB activation can be detected via luminescence assays of cellular lysate. RT-qPCR of NF-κB target gene expression can be used to quantify expression of particular genes, as well as validate functional activity of NF-κB activation. The major advantages of such a system are its simplicity and speed, which allows for high throughput screening of a range of conditions that modulate NF-κB activation. This protocol is suitable for other cell lines expressing an NF-κB::luciferase reporter, and has been demonstrated in stably transfected RAW264.7 cells18. The amount of time required to handle samples, starting from cell lysis to generating a luminescence signal, is minimal and takes the span of about an hour. Measurement of NF-κB requires only basic laboratory equipment such as opaque plates, a plate reader capable of measuring luminescence, and simple data analysis software such as a spreadsheet program.
Protocol
1. Cell Passaging and Seeding
- Maintain HeLa 57A cells in a 75 cm2 flask containing 10 mL of Dulbecco's modified Eagle's media (DMEM) supplemented with 5% heat inactivated fetal bovine serum (FBS) at 37 °C in a 5% CO2 incubator.
- Aspirate cell culture media and wash with 1 mL of 0.05% trypsin-EDTA solution. Aspirate trypsin and replace with an additional 1 mL. Place the flask in a 37 °C incubator for 4-5 min to allow cells to detach from flask.
- Add 9 mL of cell culture media and gently wash the bottom of the flask a few times to dislodge cells and form a homogenous suspension.
- Dilute HeLa 57A cells 1:6 or 1:8 in fresh media and seed into new flasks. Passage cells when they are 90% confluent or every three days and maintain cells at a minimum of 25% confluency.
- One day before cell stimulation, trypsinize HeLa 57A cells and suspend in 10 mL of growth media. Count cells in suspension using a hemocytometer and use growth media to dilute cells to a final concentration of 2.5 x 105 cells/mL in a 50 mL conical tube.
- Transfer 250 µL of cell suspension (~6.25 x 104 cells) to each well of a 48-well plate. Periodically cap and turn over the conical tube to ensure a homogenous cell suspension. Tap the plate gently on the side to ensure that the cells distribute uniformly in the wells.
- Transfer the plate to 37 °C in a 5% CO2 incubator and allow the cells to attach and grow overnight.
2. Preparation of Bacteria
- Two days before infection, streak frozen stocks of Salmonella onto LB agar plates to produce single colonies. Transfer plates to an incubator set to 37 °C and allow overnight growth.
- One day before infection, add 3 mL of lysogeny broth (LB) to sterile bacterial culture tubes, adding the appropriate antibiotics to the media.
- Using a sterile inoculation loop, pick a single colony from streaked bacterial cultures and touch the loop to the LB media. Cap the tubes after inoculating and discard the loop.
- Place the tubes in a shaking incubator set at 37 °C, 180 rpm, and allow to grow overnight.
- On the day of infection, retrieve overnight bacterial cultures from the incubator.
- Prepare tubes for subculture by adding 3 mL of fresh LB, containing antibiotics when appropriate.
- Transfer 30 µL of the overnight bacterial culture (1 in 100 dilution) to the freshly prepared media. Place the tubes in a shaking incubator set at 37 °C for 3 h.
- After the 3 h incubation, transfer 1 mL of sterile LB broth into a plastic cuvette to serve as the blank. Transfer 900 µL of LB into the other cuvettes to be used for the sample analysis.
- Transfer 100 µL of bacterial subculture into a cuvette containing 900 µL of LB and pipette up and down several times to mix. Repeat this for each bacterial suspension.
- Turn on the spectrophotometer to measure the optical density (OD) of the bacterial cultures at a wavelength of 600 nm (OD600).
- Place the blank in the spectrophotometer. Take note of the orientation, as there should be a mark towards the top indicating such.
- Close the lid and press the Blank button on the spectrophotometer to get the background absorbance.
- Replace the blank cuvette with a sample cuvette in the same orientation and press Read.
- Record the OD600 values of these samples. Multiply the value by 10 to account for the dilution factor.
- Dilute the bacterial subculture with fresh LB to achieve an absorbance value of approximately 1.0, which roughly corresponds with 1 x 109 cfu/mL. Dilute this suspension 1:10 in a cuvette and measure the absorbance — which should give a value of ~0.1.
- In a new tube, add an appropriate volume of the diluted subculture to fresh LB to achieve a suspension of 1 x 108 cfu/mL to be used as an inoculum.
- Prepare serial dilutions of the inoculum by transferring 50 µL of bacterial suspension to a tube containing 450 µL of sterile PBS (10-fold dilution) until a final dilution of approximately 102 cfu/mL is made.
- Transfer 100 µL of the two lowest dilutions (102 and 103 corresponding with 10 and 100 colony forming units, respectively) to an LB agar plate and spread the suspension with a cell spreader to obtain single colonies. Transfer these plates to a 37 °C incubator and incubate overnight.
- The following day count the colonies and calculate the bacterial concentration of the initial inoculum to determine the actual inoculum concentration.
3. Infection of Cells
NOTE: At this point, the cells should be at about 90% confluency. For HeLa 57A cells in a 48-well plate, this is approximately 1 x 105 cells per well. Cells will be infected with multiplicity of infection (MOI) of 10, or 106 cfu/well.
- Label the lid of the plate according to the infection conditions that will be used for each well, with each condition being done in triplicate.
- Add 10 µL of the inoculum to appropriate wells. Add 10 µL of sterile LB to uninfected control wells.
- To synchronize the time of infection, place the plate in a tabletop centrifuge and spin at 500 x g for 5 min, ensuring that the plate is counter balanced.
- Transfer infected cells to a 5% CO2 incubator at 37 °C for 1 h.
- Place an aliquot of cell culture media in a 37 °C water bath for use in the next step.
- One hour after time of infection, remove cell culture media from water bath and wipe the exterior with 70% ethanol. Transfer tissue culture plate to biosafety cabinet.
- Using sterile technique, aspirate media from wells and replace with 250 µL of fresh, warm cell culture media.
- Return plates to the 5% CO2 incubator at 37 °C for additional 4 h.
- Remove plates from CO2 incubator and aspirate the media from the wells. For luciferase analysis continue to step 4.1. For RNA isolation proceed to step 5.1.
4. Luciferase Analysis
- Add 100 µL of 1x cell lysis buffer to the wells. The buffer may need to first be diluted to a working concentration, depending on the reagent.
- Transfer the plate to a -80 °C freezer and incubate for at least 30 min to ensure efficient cell lysis.
- Place the plate containing frozen cell lysate on a bench to thaw and prepare luciferase substrate reagents according to manufacturer recommendations.
- Allow luciferase substrate reagents to equilibrate to room temperature.
- Turn on the plate reader and open the corresponding reader program. Set the machine to measure luminescence.
- Transfer 10 µL of each sample to a well of an opaque 96-well plate.
- Add 50 µL of the Luciferase Assay Reagent using a multichannel pipette to each well of the opaque plate.
- Gently tap the plate on the side to mix wells and ensure that the bottom surface is covered with liquid. Place the plate in a plate reader and initiate reading.
- Copy the luminescence values into a spreadsheet program and plot the results.
5. RNA Isolation
- Add 500 µL of guanidium thiocyanate to each well and pipette up and down several times to ensure complete lysis.
- Transfer the contents to a labeled microcentrifuge tube. Maintain samples on ice as much as possible to maintain RNA quality.
- Set a tabletop centrifuge to 4 °C and maintain the centrifuge at this temperature for remainder of protocol.
- To each sample tube, add 100 µL of chloroform, cap tightly, and shake for 15 s. Incubate at room temperature for 10 min.
- Centrifuge at 12,000 x g for 15 min at 4 °C. During this time, prepare for the next step of RNA purification by labeling new tubes.
- Transfer the upper aqueous phase to the new tube containing 250 µL of isopropanol, being careful not to disturb the middle or lower layers.
- Store unused product in a -80 °C freezer, which can also be used for protein analysis. The residual aqueous layer may also serve as a backup if RNA is needed later.
- Vortex the mixture of aqueous layer and isopropanol and allow samples to sit at room temperature for 10 min.
- Centrifuge the samples at 12,000 x g for 10 min at 4 °C.
- Remove tubes from centrifuge. RNA precipitate forms a white pellet on the side and bottom of the tube. Open the tubes and carefully remove the supernatant by pouring out into a waste container.
- Wash the RNA pellet by adding 500 µL of 75% ethanol and vortex.
- Centrifuge at 8,000 x g for 5 min at 4 °C.
- Remove the supernatant by pouring out and again wash the pellet with 75% ethanol.
- Centrifuge at 8,000 x g for 5 min at 4 °C.
- Using a small volume pipette (e.g., 10-200 µL volume), aspirate as much of the supernatant as possible, being careful not to push any liquid back into the tube or dislodge the pellet with the pipette tip. If the pellet becomes dislodged, centrifuge briefly and again try to remove supernatant.
- Air-dry the RNA pellets. If the pellets are visible for any sample, periodically (every 5 minutes) check on them to see if they change from white to clear, indicating that they are dry. Once the pellets begin turning clear, add 20 µL of ultrapure, RNase free water to dissolve the RNA.
- If pellets were not initially visible for any samples, simply add ultrapure water 5 min after removing supernatant.
- To increase the solubility, pass the solution a few times through a pipette tip and incubate for 10 min at 55-60 °C.
6. DNase Treatment of RNA
- To maintain RNA integrity, store RNA samples on ice unless otherwise noted. At this time, the 10x DNase buffer can be removed from the freezer and allowed to thaw on ice.
- Prepare the spectrophotometer for sample analysis by cleaning all sample analysis surfaces with a lint free cloth.
- Take a background measurement prior to analysis. To do so, load the sensor with 1.5 µL of the same ultrapure water used to dissolve the RNA pellets. Press the Read Blank button to generate a background reading.
- Use the lint free cloth to wipe the instrument's sample loading surface. Repeat this step after each sample reading.
- Load 1.5 µL of resuspended RNA to the sample holder and select Read Sample. Repeat until all samples have been read.
- Prepare DNase master mix by combining 2.4 µL of 10x DNase buffer and 1 µL of DNase, per sample, in a 1.5 mL microcentrifuge tube. Prepare the master mix for two extra volumes.
- Mix the master mix by flicking the tube several times. Briefly centrifuge the tube to collect contents at the bottom of the tube. Add 3.4 µL of mastermix to 20 µL of RNA sample. Mix contents by flicking and centrifuge briefly, as before.
- Transfer the samples to a heat block set at 37 °C for 20 min. Remove the DNase inactivation reagent from the freezer and thaw at room temperature.
- 20 min after placing samples on the heat block, transfer the samples to a tube rack placed on the work bench.
- Gently vortex the contents of the DNase inactivation reagent. Transfer 2.6 µL of DNase inactivation reagent to the tubes containing RNA and then flick the tubes to create a homogenous mixture.
- Centrifuge the samples at 12,000 x g for 1 min.
- Carefully pipette the supernatant to a new, labeled tube.
7. Reverse Transcription of mRNA to cDNA
- Prepare a master mix depending on the amount of samples plus two extra to account for loss through pipetting (Table 1). Use 1 µg of RNA and add H2O to a volume of 50 µL total.
| Reagent | Volume (μL) | Final Concentration |
| MgCl2 (25mM) | 3.5 | 1.75 mM |
| Reverse transcription buffer (10x) | 5 | 1x |
| dNTP mix (10 μM, each) | 2.5 | 500 nm |
| random hexamer (100 μM) | 1.25 | 2.5 μM |
| Multiscribe reverse transcriptase (50 U/μL) | 1 | 1 U/μL |
| RNase inhibitor (20 U/μL) | 1.25 | 1.25 U/μL |
| RNA (1 μg) | 20 ng/uL | |
| H2O | top up to 50 |
Table 1: Components and recipe for reverse transcription master mix.
- Cap the samples tightly and label the PCR tubes on the side as labels on the lid may be removed from the heated lid of the thermocycler later. Mix by vortexing.
- Briefly centrifuge the PCR tubes to collect samples to the bottom of the tube.
- Place the PCR tubes in a thermocycler and run the samples under the following settings:
25 °C for 10 min, 48 °C for 30 min, 95 °C for 5 min, and then hold at 10 °C. - Transfer tubes containing newly synthesized cDNA to a -20 °C freezer or use immediately in qPCR analysis.
8. Preparing and Loading Plate for RT-qPCR Analysis
- Before starting, plan the setup of the 384-well qPCR plate for sample analysis.
- Thaw primers and cDNA on ice.
- Prepare a master mix (see Table 2), plus approximately 10% extra to account for loss through pipetting.
| Reagent | Volume (µL) |
| 10 µM F primer | 1 |
| 10 µM R primer | 1 |
| Ultrapure H2O | 4 |
| 2x SYBR green | 10 |
Table 2: Components and recipe for qPCR master mix.
- Vortex the master mix and dispense 8 µL into the wells of the 384-well plate using a repeater pipette. Samples will be analyzed in duplicate for each primer and cDNA sample combination.
- Using a P10 (or smaller) pipette, transfer 2 µL of cDNA to duplicate wells for each primer set to be analyzed. Replace tips after each well to avoid cross contamination.
- Seal the plate by carefully applying the adhesive film to the surface, ensuring that all wells are covered. Press the film using a sealing paddle or roller to seal firmly.
- Place the plate in a centrifuge, with an empty plate as a counterbalance. Centrifuge the plate at 500 x g for 5 min.
9. Running the Thermocycler for qPCR Analysis
- Power on the computer and real-time PCR instrument.
- Open the RT-qPCR software and Select New Experiment.
- Under the Setup tab, select Experiment Properties, where the parameters for the run can be set.
- Define the Experiment Name, which will set the file name and settings used to store results.
- For the Instrument Selection, select the currently connected instrument that will be running the analysis. Select Comparative CT (ΔΔCt) run method.
- Select SYBR Green Reagents as the fluorescent DNA dye to be used and Standard as the ramp speed.
- Ensure that the box next to Include Melt Curve is checked.
- Under the Define tab, assign targets (i.e., genes to be amplified), and samples (i.e., experimental conditions).
- Select the Assign tab. Label the wells with the appropriate targets and samples, as they correspond to the loading scheme of the 384-well plate.
- Select Run Method. Use the parameters for analysis listed in (Table 3).
| Hold Stage | PCR Stage | Melt Curve Stage | |||||
| Step 1 | Step 2 | Step 1 | Step 2 | Step 1 | Step 2 | Step 3 | |
| Temp | 50 ℃ | 95 ℃ | 95 ℃ | 60 ℃ | 95 ℃ | 60 ℃ | 95 ℃ |
| Time | 2:00 | 10:00 | 0:15 | 1:00 | 0:15 | 1:00 | 0:15 |
| Data Collection | yes | yes | |||||
| Number of cycles | 1x | 40x | 1x | ||||
Table 3: Cycle parameters for thermocycler.
- Place the 384-well plate in the thermocycler and start the analysis.
10. Analysis of qPCR Results with the Delta-delta Ct Method (2-ΔΔCt)
- Analyze results generated from the qPCR reaction for errors that may interfere with downstream analysis. Many errors will be flagged automatically by the system.
- For properly constructed primers, the melt curves should only have one peak. Exclude any wells containing a melt curve with more than one peak from further analysis.
- Export the Ct values to a spreadsheet program to analyze the data using the ΔΔCt method. A suggested format is provided (Table 4). The last column is the fold expression change of the sample, relative to the control samples.
| Housekeeping gene (GAPDH) | Gene of interest (IL6) | ||||||||||
| Ct1 | Ct2 | Ave Ct | Ct1 | Ct2 | Ave Ct | ΔCt | Ave ΔCt ctrls | ΔΔCt | 2^-(ΔΔCt) | Geomean | |
| Control 1 | 15.33 | 15.37 | 15.35 | 26.81 | 26.91 | 26.86 | 11.51 | 10.51 | 1.00 | 0.50 | 1.00 |
| Control 2 | 16.83 | 16.77 | 16.80 | 26.89 | 26.92 | 26.91 | 10.11 | 10.51 | -0.41 | 1.33 | |
| Control 3 | 17.56 | 17.53 | 17.54 | 27.38 | 27.56 | 27.47 | 9.93 | 10.51 | -0.59 | 1.50 | |
| Sl1344 1 | 15.50 | 15.41 | 15.45 | 22.15 | 22.13 | 22.14 | 6.69 | 10.51 | -3.83 | 14.21 | 13.23 |
| SL1344 2 | 16.02 | 15.98 | 16.00 | 23.01 | 22.96 | 22.98 | 6.98 | 10.51 | -3.53 | 11.57 | |
| SL1344 3 | 17.27 | 17.30 | 17.28 | 23.99 | 23.98 | 23.98 | 6.70 | 10.51 | -3.82 | 14.09 | |
| sipA sopB sopE2 1 | 15.38 | 15.41 | 15.39 | 23.31 | 23.09 | 23.20 | 7.80 | 10.51 | -2.71 | 6.56 | 7.29 |
| sipA sopB sopE2 2 | 16.01 | 16.05 | 16.03 | 23.89 | 23.92 | 23.91 | 7.88 | 10.51 | -2.64 | 6.23 | |
| sipA sopB sopE2 3 | 16.78 | 16.78 | 16.78 | 24.02 | 24.06 | 24.04 | 7.27 | 10.51 | -3.25 | 9.49 | |
| sipA sopB sopE2 sopE 1 | 15.52 | 15.60 | 15.56 | 27.04 | 27.03 | 27.03 | 11.47 | 10.51 | 0.96 | 0.51 | 0.79 |
| sipA sopB sopE2 sopE 2 | 15.56 | 15.59 | 15.57 | 26.37 | 26.42 | 26.39 | 10.82 | 10.51 | 0.31 | 0.81 | |
| sipA sopB sopE2 sopE 3 | 15.91 | 15.92 | 15.91 | 26.24 | 26.12 | 26.18 | 10.27 | 10.51 | -0.25 | 1.19 | |
Table 4: Format for analyzing qPCR data.
Representative Results
The assay described here focuses on activation of the transcription factor NF-κB using a NF-κB-dependent luciferase reporter that is stably transfected into a line of HeLa cells. Activated NF-κB translocates to the nucleus where it binds κB binding sites of target genes, including the pro-inflammatory cytokines IL6 and IL23. A general overview of NF-κB activation is depicted in Figure 1. The gram-negative bacterium Salmonella enterica serovar Typhimurium was used in this study as an activator of NF-κB and subsequent expression of IL6 (Figure 2). One of the main virulence factors required for pathogenesis is the Type III Secretion System-1 (T3SS-1) that allows S. Typhimurium to infect cells and to induce NF-κB activation. The function of the T3SS-1 is to deliver bacterial proteins, termed effectors, into host cells. Here effector proteins target numerous cellular signaling pathways to mediate invasion of host epithelial cells. The NF-κB activation induced by S. Typhimurium is mostly dependent on the T3SS-1 effector proteins SopE, SipA, SopB and SopE218. Here, HeLa 57A cells were infected with the wild type S. Typhimurium strain SL1344 and two mutant strains lacking either three (SipA, SopB, SopE2) or four (SipA, SopB, SopE2, SopE) effector proteins. Figure 2 is a representative experiment of NF-κB-dependent luciferase activation (Figure 2A) and IL6 gene expression (Figure 2B) in HeLa 57A cells infected with the S. Typhimurium strains. Infection with the wild type SL1344 strain induces a strong luciferase signal which is decreased in cells infected with the SipA, SopB, SopE2 triple mutant (SopE-dependent response), and reduced to control levels with the SipA, SopB, SopE2, SopE quadruple mutant. The relative luminescence units (RLU) correlates with the IL6 expression levels (Figure 2). Figure 3 represents the results generated from RT-qPCR analysis.
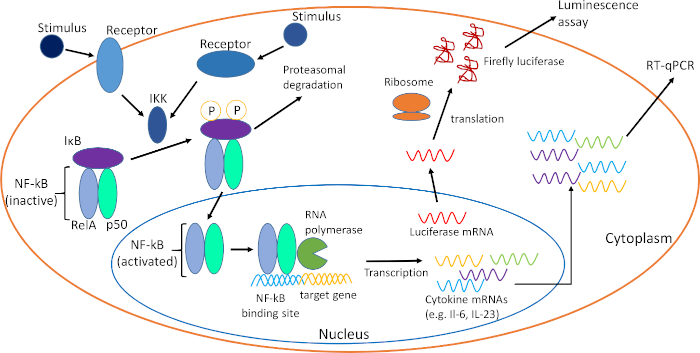
Figure 1: Schematic of NF-κB activation and downstream readouts. Outlined above, NF-κB is retained in the cytosol in an inactive state by the inhibitory protein IκBα. Both internal and external stimuli can contribute to activation of IKK, which phosphorylates IκBα and causes its subsequent proteosomal degradation. Degradation of IκBα reveals the nuclear localization signal of NF-κB, which promotes translocation. In the nucleus, activated NF-κB binds to κB binding sites of target genes, promoting their transcription and expression. Target genes, such as the cytokines IL6 and IL23, are expressed and quantified via RT-qPCR. In HeLa 57A cells luciferase is expressed following NF-κB activation, which can be quantified via luminescence assays. Please click here to view a larger version of this figure.
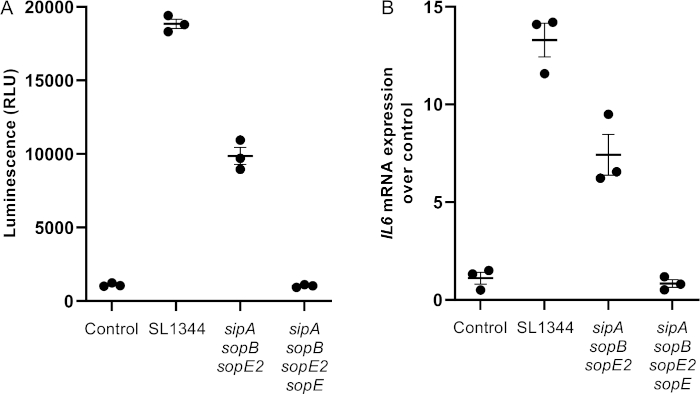
Figure 2: Representative data of luminescence assay and RT-qPCR analysis following S. Typhimurium infection of HeLa 57A cells. 1 x 105 HeLa 57A cells were infected with 106 cfu (MOI = 10) of log phase S. Typhimurium wild type SL1344, or the mutant strains lacking SipA, SopB and SopE2, and SipA, SopB, SopE2 and SopE. After 1 h, media was replaced, and incubation continued for an additional four hours. (A) Cells were processed for expression of luciferase via luminescence assays. (B) Cells were extracted of RNA, which was used for RT-qPCR analysis. Relative expression of target genes using the delta-delta Ct method (2–ΔΔCt) is shown. Mean and standard deviation of triplicate wells are shown. Please click here to view a larger version of this figure.
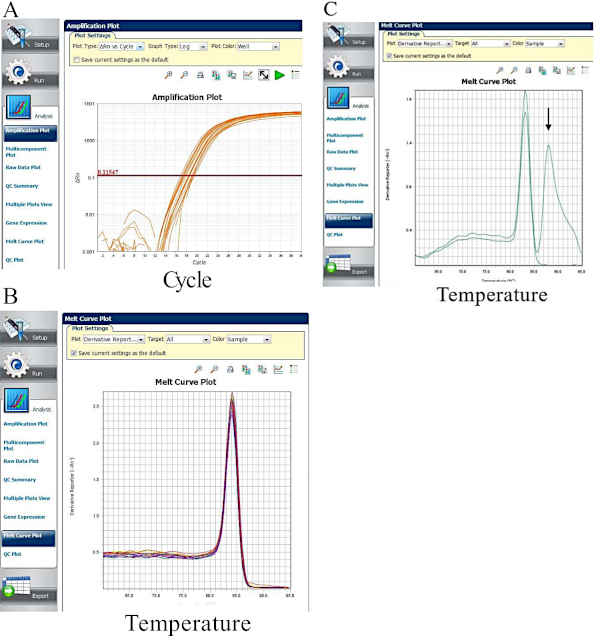
Figure 3: Summary results generated from RT-qPCR analysis. (A) An amplification plot is shown for the wells amplifying GAPDH from HeLa 57A cells infected with S. Typhimurium. (B) Melt curve plot of wells containing GAPDH primers shows a single melt peak corresponding to approximately 84 °C. (C) Duplicate wells containing primers for IL-6. One of the wells displays a second melt peak, as denoted by a black arrow, and should be excluded from analysis. Please click here to view a larger version of this figure.
Discussion
The major contribution of the protocol described is that it provides a fast and easy method to detect NF-κB activation in cells, which allows for high throughput analysis of multiple stimulatory conditions or drugs affecting NF-κB activation. Here, we describe a protocol for NF-κB activation in Salmonella-infected HeLa cells. These cells can be used for infection with other pathogens as well to study the impact of bacterial infection on NF-κB activation. In addition, NF-κB-dependent luciferase activation in HeLa 57A cells can be used to screen for activators or inhibitors of signaling pathways leading to NF-κB activation. Here, we seed HeLa 57A cells in 48-well plates, but these experiments can be scaled up to 96- and even 384-well plates to allow for more samples per experiment. HeLa 57A cells constitutively express LacZ, which can be measured as an internal control for cell number and viability using β-gal assays. The protocol described here is applicable for use in cell lines either stably or transiently transfected with an NF-κB::luciferase reporter construct. The RAW264.7 macrophage cell line has already been used for such a purpose18. Importantly, this method does not distinguish between activation of the different homo/heterodimers of the five distinct NF-κB subunits. The different NF-κB homo- heterodimer complexes have different affinities for promoter sequences, as well as differing transcriptional consequences21. It is possible that activation of a specific NF-κB dimer is missed using the reporter described here due to low affinity binding to the κB binding sites from the immunoglobulin κ-chain promoter region.
It is important to note that the conditions suitable for stimulating one cell line may not be directly applicable to another. Therefore, it is highly recommended that the assay conditions be optimized in each assay system. Time of infection, MOI, duration of drug treatment, drug dose, and seeding conditions are all important considerations to take into account when assessing NF-κB activation. For example, RAW264.7 macrophages are more sensitive to Salmonella infection requiring different conditions to produce a similar luminescence signal as seen with HeLa 57A cells18.
During luminescence measurements, it is not uncommon for edge wells to have considerably different values than the other wells that were similarly stimulated. This is typically due to higher evaporation rates of the edge wells on the plate leading to increased drug concentration. This may be addressed by adding serum free media to the space between the wells, for plates that allow it, altering lid/plate combinations to reduce evaporation, or omit edge wells completely if issues persist.
Expressed in mammalian cells, firefly luciferase has a half-life of several hours and is generally regarded as stable for the purposes of most reporter assays22. Longer treatment durations than those described here may be reliably carried out. However, the luciferase assay system used here produces a stable signal only for the first minute and quickly deteriorates after that. Therefore, it is highly important that the luminescence measure be made quickly after mixing cell lysate and substrate.
NF-κB is a transcription factor for a large array of genes including cytokines and chemokines (i.e., Il6 and Il23). Measuring NF-κB-dependent luciferase activity does not necessarily mean these cytokines and chemokines are expressed. This method can complement existing molecular quantification techniques by serving as a potential first screen for the characterization of conditions affecting NF-κB activity, which can then be functionally validated through RT-qPCR. Functional validation of NF-κB activation can also be performed via western blot analysis or functional assays specific to a protein of interest in cases where mRNA quantification may not be appropriate. This is the case for interleukin-beta (Il1b), a gene whose transcription is induced by NF-κB binding, but the protein product requires post translational modification to form the mature product23,24.
The specific RNA isolation protocol described here is not the only one that may be employed for use in the RT-qPCR analysis, and other preferred methods, such as commercially available kits, can instead be used. It is important to note that the quality of the RNA is very important to the process and so RNA should be kept on ice as much as possible to prevent degradation. It is important to run duplicate reactions in the RT-qPCR reactions to ascertain that the PCR reactions are consistent, as the pipetting of such small volumes when loading the 384-well plate can often be cause for error. Ct values of the housekeeping gene should be consistent between samples, and deviations from this may be indicative of inefficient cleaning steps or discrepancies in RNA concentrations during reverse transcription that may affect the final results. It is also important to check the melt curve after each qPCR experiment. The presence of one peak suggests that the qPCR primers are amplifying a single gene. Multiple curves suggest that there is off-target gene amplification or presence of primer dimers. In either case, multiple peaks present in the melt curve suggests that the primers should be redesigned to ensure specificity. With so much room for variability, exercise and refinement of good technique at every step are essential for generating accurate and reproducible results.
Declarações
The authors have nothing to disclose.
Acknowledgements
Research in the Keestra-Gounder lab is supported by grants from the NIAID of the NIH under Award Number R21AI122092 and from the American Diabetes Association under Award Number 1-18-JDF-035.
Materials
| Adhesive film | VWR International | 60941-070 | |
| chloroform | Fisher Bioreagents | C298-500 | |
| DMEM | Thermo Fisher | 11665092 | |
| DNAse treatment kit | Qiagen | 79254 | |
| dNTPs | Promega | U1511 | |
| ethanol | Fisher Bioreagents | BP2818100 | molecular grade |
| FBS | Sigma-Aldrich | F0926 | |
| HeLa 57A cells | Ref # 15 | ||
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | 4368814 | |
| isopropanol | Fisher Bioreagents | BP26181 | |
| Kanamycin | Fisher Bioreagents | BP906-5 | |
| LB agar | Fisher Bioreagents | BP1425-500 | |
| Lysogeny broth | Fisher Bioreagents | BP1426-500 | |
| MgCl2 | Fisher Chemical | ||
| NanoDrop ND-1000 | Thermo Scientific | spectrophotometer | |
| promega luciferase assay system | Promega | E1501 | Cell lysis buffer & luciferin substrate |
| Random Hexamers | Thermo Scientific | SO142 | |
| Real-time GAPDH forward primer | 5'-CCAGGAAATGAGCTTGAC AAAGT-3' |
||
| Real-time GAPDH reverse primer | 5-'CCCACTCCTCCACCT TTGAC-3' |
||
| Real-time IL-23 forward primer | 5-'GAGCCTTCTCTGCTCCC TGAT-3' |
||
| Real-time IL-23 reverse primer | 5'-AGTTGGCTGAGGCCCAGTAG-3' | ||
| Real-time IL-6 forward primer | 5'-GTAGCCGCCCCACACAGA-3' | ||
| Real-time IL-6 reverse primer | 5'-CATGTCTCCTTTCTCAGG GCTG-3' |
||
| Reverse Transcriptase | Applied Biosystems | 4308228 | |
| RNAse inhibitor | Thermo Scientific | EO0381 | |
| RT buffer | Promega | A3561 | |
| SL1344 | Ref # 17 | ||
| SL1344 ΔsipA sopB::MudJ sopE2::pSB1039 | Ref # 18 | ||
| SL1344 ΔsopE ΔsipA sopB::MudJ sopE2::pSB1039 | Ref # 18 | ||
| SYBR green | Applied Biosystems | 4309155 | 2x mastermix |
| Tri-reagent | Molecular Research Center | TR 118 | guanidine thiocyanate |
| Trypsin -EDTA | Thermo Fisher | 25300054 | 0.05% Trypsin-EDTA |
| ultrapure water | Fisher Bioreagents | BP248450 | |
| Well plate for PCR | VWR International | 89218-294 | 384-well plate |
Referências
- Liu, T., Zhang, L., Joo, D., Sun, S. C. NF-kappaB signaling in inflammation. Signal Transduction and Targeted Therapy. 2, (2017).
- Piva, R., Belardo, G., Santoro, M. G. NF-kappaB: a stress-regulated switch for cell survival. Antioxidants & Redox Signaling. 8 (3-4), 478-486 (2006).
- Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harbor Perspectives in Biology. 1 (6), a001651 (2009).
- Hargett, D., Rice, S., Bachenheimer, S. L. Herpes simplex virus type 1 ICP27-dependent activation of NF-kappaB. Journal of Virology. 80 (21), 10565-10578 (2006).
- Zaragoza, C., et al. Viral protease cleavage of inhibitor of kappaBalpha triggers host cell apoptosis. Proceedings of the National Academy of Sciences United States of America. 103 (50), 19051-19056 (2006).
- Gao, X., et al. Bacterial effector binding to ribosomal protein s3 subverts NF-kappaB function. PLOS Pathogens. 5 (12), e1000708 (2009).
- Kravchenko, V. V., et al. Modulation of gene expression via disruption of NF-kappaB signaling by a bacterial small molecule. Science. 321 (5886), 259-263 (2008).
- Kawai, T., Akira, S. Signaling to NF-kappaB by Toll-like receptors. Trends in Molecular Medicine. 13 (11), 460-469 (2007).
- Pinaud, L., Sansonetti, P. J., Phalipon, A. Host Cell Targeting by Enteropathogenic Bacteria T3SS Effectors. Trends in Microbiology. 26 (4), 266-283 (2018).
- Karin, M. How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene. , (1999).
- Berti, R., et al. Quantitative real-time RT-PCR analysis of inflammatory gene expression associated with ischemia-reperfusion brain injury. Journal of Cerebral Blood Flow & Metabolism. 22 (9), 1068-1079 (2002).
- Fried, M., Crothers, D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Research. 9 (23), 6505-6525 (1981).
- Holden, N. S., Tacon, C. E. Principles and problems of the electrophoretic mobility shift assay. Journal of Pharmacological and Toxicological Methods. 63 (1), 7-14 (2011).
- Renard, P., et al. Development of a sensitive multi-well colorimetric assay for active NFkappaB. Nucleic Acids Research. 29 (4), E21 (2001).
- Nowak, D. E., Tian, B., Brasier, A. R. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 39 (5), 715-725 (2005).
- Ernst, O., Vayttaden, S. J., Fraser, I. D. C. Measurement of NF-kappaB Activation in TLR-Activated Macrophages. Methods in Molecular Biology. 1714, 67-78 (2018).
- Rodriguez, M. S., Thompson, J., Hay, R. T., Dargemont, C. Nuclear retention of IkappaBalpha protects it from signal-induced degradation and inhibits nuclear factor kappaB transcriptional activation. Journal of Biological Chemistry. 274 (13), 9108-9115 (1999).
- Mendez, J. M., Kolora, L. D., Lemon, J. S., Dupree, S. L., Keestra-Gounder, A. M. Activation of the endoplasmic reticulum stress response impacts the NOD1 signaling pathway. Infection and Immunity. , (2019).
- Hoiseth, S. K., Stocker, B. A. D. Aromatic-Dependent Salmonella-Typhimurium Are Non-Virulent and Effective as Live Vaccines. Nature. 291 (5812), 238-239 (1981).
- Keestra, A. M., et al. Manipulation of small Rho GTPases is a pathogen-induced process detected by NOD1. Nature. 496 (7444), 233-237 (2013).
- Wong, D., et al. Extensive characterization of NF-kappaB binding uncovers non-canonical motifs and advances the interpretation of genetic functional traits. Genome Biology. 12 (7), R70 (2011).
- Gupta, R., et al. Firefly luciferase mutants as sensors of proteome stress. Nature Methods. 8 (10), 879-884 (2011).
- Cogswell, J. P., et al. NF-kappa B regulates IL-1 beta transcription through a consensus NF-kappa B binding site and a nonconsensus CRE-like site. Journal of Immunology. 153 (2), 712-723 (1994).
- Thornberry, N. A., et al. A Novel Heterodimeric Cysteine Protease Is Required for Interleukin-1-Beta Processing in Monocytes. Nature. 356 (6372), 768-774 (1992).

