Qualitative and Comparative Cortical Activity Data Analyses from a Functional Near-Infrared Spectroscopy Experiment Applying Block Design
Summary
We describe the analysis of continuous-wave functional near-infrared spectroscopy experiment using a block design with a sensorimotor task. To increase the reliability of the data analysis, we used the qualitative general linear model-based statistical parametric mapping and the comparative hierarchical mixed models for multi-channels.
Abstract
Neuroimaging studies play a pivotal role in the evaluation of pre- vs. post-interventional neurological conditions such as in rehabilitation and surgical treatment. Among the many neuroimaging technologies used to measure brain activity, functional near-infrared spectroscopy (fNIRS) enables the evaluation of dynamic cortical activities by measuring the local hemoglobin levels similar to functional magnetic resonance imaging (fMRI). Also, due to lesser physical restriction in fNIRS, multiple variants of sensorimotor tasks can be evaluated. Many laboratories have developed several methods for fNIRS data analysis; however, despite the fact that the general principles are the same, there is no universally standardized method. Here, we present the qualitative and comparative analytic methods of data obtained from a multi-channel fNIRS experiment using a block design. For qualitative analysis, we used a software for NIRS as a mass-univariate approach based on the generalized linear model. The NIRS-SPM analysis shows qualitative results for each session by visualizing the activated area during the task. In addition, the non-invasive three-dimensional digitizer can be used to estimate the fNIRS channel locations relative to the brain. To corroborate the NIRS-SPM findings, the amplitude of the changes in hemoglobin levels induced by the sensorimotor task can be statistically analyzed by comparing the data obtained from two different sessions (before and after intervention) of the same study subject using a multi-channel hierarchical mixed model. Our methods can be used to measure the pre- vs. post-intervention analysis in a variety of neurological disorders such as movement disorders, cerebrovascular diseases, and neuropsychiatric disorders.
Introduction
Neurorehabilitation plays an important role in the functional recovery following sensorimotor disturbance. To clarify the mechanisms of neuroplasticity-associated functional recovery, various neuroimaging technologies have been used, such as functional magnetic resonance imaging (fMRI), positron emission tomography (PET), electroencephalography (EEG), and functional near-infrared spectroscopy (fNIRS). Different imaging modalities have different advantages and disadvantages. Although the fMRI is the most typical device, it is affected by magnetic fields, has a high cost, high physical restriction, and limited sensorimotor tasks1,2,3,4. The fNIRS device stands out as a noninvasive optical neuroimaging and has a relatively lower spatial resolution, but it has a better temporal resolution than fMRI4. fNIRS is suitable when verifying treatment effects because it compares the pre- versus post-intervention effects, has dynamic motor tasks, is portable, and functions more in natural environments than fMRI1,2,4. NIRS has been reported to be more suitable in the fields of cerebrovascular disease, epileptic disorders, severe brain injury, Parkinson's disease, and cognitive impairment1,5. With regard to sensorimotor tasks, it is widely used in gait and standing balance6,7,8, upper limb function (hand grasping, finger tapping)8,9, complex motor skill training10,11, robotics12,13,14,15, and brain-computer interface16,17,18. The fNIRS is based on the principles of optical neuroimaging and neurovascular coupling, which measure cortical metabolic activity, increased blood flow, and consequently cortical activity as secondary signals19. fNIRS signals have been reported to have strong correlations with signals of blood oxygen level-dependent fMRI20. A continuous-wave fNIRS uses the modified Beer-Lambert law to determine the changes in oxygenated hemoglobin (HbO2) and deoxygenated hemoglobin (HHb) cortical concentration levels based on measured changes in broadband near-infrared light attenuation21,22. Because it was not possible to measure the differential path-length factor (DPF) using the continuous-wave NIRS system, we assumed that the DPF was constant and that hemoglobin signal changes were denoted in arbitrary units of millimole-millimeter (mM x mm)2,18.
The fNIRS experiments need to select the most adequate methods including the probe settings, the experiment designs, and the analysis methods. Regarding the probe setting, the international 10-20 method used in EEG measurement is the setting standard used by many researchers in neuroimaging. In recent years, coordinate settings based on the standard brain on the basis of Montreal Neurological Institute (MNI) coordinates have been used. The experiment uses a block design, generally used for sensorimotor tasks, and an event-related design. This is a method of comparing changes in hemoglobin concentration at rest and during tasks; HbO2 concentration levels increase and HHb concentration levels decrease with changes in cerebral blood flow associated with task-dependent cortical activity. Although there are various analysis methods, the NIRS-SPM free software enables an analysis similar to the statistical parametric mapping (SPM) of fMRI. The treatment of NIRS data uses a mass-univariate approach based on the general linear model (GLM). When performing task-dependent brain activity analysis, the fNIRS measurements can be affected by evoked or non-evoked neuronal activity and systemic physiological interferences (heart rate, blood pressure, breathing rate, and autonomic nervous system activity) in the cerebral and extracerebral compartment23. Therefore, pre-analysis processing, filtering, wavelet conversion, and principal component analysis are useful23. Regarding filtering and artifacts of the data processing using the NIRS-SPM, low-pass filtering9 and the wavelet minimum description length (Wavelet-MDL)24 detrending were used to overcome the motion or other sources of noise/artifact. For details of this analytic method, refer to the report of Ye et al.25. Although there are reports using only SPM, it is only a qualitative index by image analysis, and due to the low spatial resolution of NIRS, extreme caution is required for group analysis. Moreover, when the DPF is constant, numerical comparisons between channels and individuals should not be performed, but the difference in the changes in each channel can be verified. Based on the above conditions, in order to supplement the NIRS-SPM group analysis results, we used the original analysis method for multi-channel analysis after improving the accuracy of spatial registration. This multi-channel analysis compared the amplitude of the change in HbO2 and HHb levels between the rest and on-task periods at each channel before and immediately after treatment using hierarchical mixed models with fixed interventions (before or after), fixed periods (rest or on-task), and random individual effects.
In this way, there are several fNIRS measurement and analysis methods; however, no standard method has been established. In this paper, we introduce our methods, qualitative GLM-based statistical parametric mapping and the comparative multi-level hierarchical mixed model, to analyze data obtained from a multi-channel fNIRS experiment of pre- vs. post-intervention using a block design with sensorimotor tasks.
Protocol
This study was approved by the institutional review board (IRB) of the Fukuoka University, Japan (IRB No. 2017M017). Prior to participation, all patients provided written informed consent.
1. Preparation of the fNIRS experiment
NOTE: A multi-channel continuous-wave laser-based NIRS system for this experiment was used. The wavelengths of the near-infrared light were 780 nm, 805 nm, and 830 nm, and the sampling rate was set at 7.8 Hz. The time and spatial resolution (distances between the light emitter and detector probe) were 0.13 s and 3.0 cm, respectively.
- Set the fNIRS device in a dark noiseless place. Conduct experiments at room temperature. Start the fNIRS instrument 30 min before the experiment.
NOTE: fNIRS systems are used under controlled temperature and humidity conditions to ensure stable operation of the equipment26. - Use a whole head caps for fNIRS recording and attach the head cap on the subject's head such that the position corresponding to the central (Cz) of international 10-20 system is located at the holder No. 245 of the head cap. (Figure 1).
- Attach the marking sticker to reference locational points: the nasion (Nz), right external auditory meatus (AR), and left external auditory meatus (AL).
NOTE: Because the three-dimensional (3D) coordinates are read around the positions of the Nz, AR, AL marking sticker and Cz holder, it is necessary to attach the marking sticker before taking a picture. - After the calibration of a high-resolution digital camera for spatial registration, take pictures of the subject's head with the probe location while showing the reference points (Cz, Nz, AR, and LR) from 15 perspectives.
NOTE: Please take a picture before placing the probe. If a picture is taken after placing the probe, the marker landmark may be hidden by the probe and wiring cord. As recommended by the manufacturer, after taking 12 pictures 30° diagonally forward to the right of the subject, take three or more pictures slightly above so that the Cz (holder No. 245) appears in the picture. This is because it is easy to make it three-dimensional when capturing a total of 15 or more shots. - Carefully separate the subject's hair that interferes with the optode using a light-emitting diode (LED)-lit plastic rod to attach the probe. Arrange the probe so that the optodes are attached at a minimal distance from the scalp surface and in contact with the scalp.
NOTE: Check carefully whether there is any pressure or discomfort for the patient due to the attachment of the optodes, because of increasing strength of the systemic confounders associated with autonomic nervous system activation23. - Arrange the 48-channel system with 32 optodes (16 light sources and 16 detectors; 4 x 4 array for each hemisphere) to a head cap bilaterally over the frontal and parietal areas as regions of interest (Figure 2).
- Start and use the 3D-digitizer software to determine the spatial registration.
- After scanning the picture data of an entire head, determine the spatial coordinate of each patient by auto-measuring and save as the Origin and the Others file (*CSV file).
NOTE: If the coordinate points could not be detected from the images using the automatic measurement, input the adjustment manually.
2. Run the experiment
- Select a block design for the experiment, and the task can be any movement of interest for the study such as hand opening/closing, finger tapping, etc. In our previous study, the task was the robot-assisted elbow movements15.
NOTE: Each cycle is made of three blocks (15 s of rest – 15 s of task – 15 s of rest), and each patient completes seven cycles in each session. - Make the participant wait in a comfortable position until the starting signal. Instruct the subject to close their eyes during the rest and the task.
- Give start and stop cues (i.e., "Repeat flexion and extension of the elbow", "Stop and relax").
NOTE: Do not speak during measurements. Carefully check for artifacts on the monitor screen during measurements. - Perform the block design task in the same posture. Upright posture with standing or sitting position is desirable not to distort the headset.
NOTE: If the patient feels uncomfortable after wearing the head holder for an extended period of time, remove or loosen the probe during robot-assisted exercise. - After completing the NIRS measurement, remove the head holder and marking sticker to end the experiment.
NOTE: Carefully check for skin damage to the scalp due to prolonged wearing.
3. Qualitative GLM analysis using NIRS-SPM software
- Start the NIRS-SPM on the MATLAB software. Convert the data file related to the change in HbO2 and HHb concentration acquired from the NIRS device to the file format for NIRS-SPM analysis.
- Choose the using NIRS system option from the pop-up menu. Select the load button and choose the convert HbO2 and HHb concentration change options.
- Detect the spatial registration of NIRS channel location. Select the Stand-alone checkbox and then select the With 3D Digitizer checkbox.
- Within From Real Coordinates to MNI Space, use the dialog box to choose _origin.CSV referring to the coordinate reference point file, and _others. CSV referring to the coordinate probes/channels file.
- Select the Registration button. Choose the points to proceed to spatial estimation, and click on the OK button. Click on the Project MNI coordinate to Rendered Brain button.
NOTE: The spatial position of the NIRS channel locations is estimated on the basis of Montreal Neurological Institute (MNI) brain template. - Select the Dorsal View option, and click on the Save button.
- In the Specify the 1st Level section, select the NIRS data filename and SPM directory. Select the hemoglobin checkbox; HbO2 or HHb. Highlight the Specify design' option and select the Sec option. Highlight the Number of Condition/Trials options and enter the number 7.
- Highlight the Vector of Onset and Duration[s] options and enter a vector of onset multiplied by the duration of the experimental conditions as follows.
NOTE: In this case, the vector of onset times should be specified as [15:45:285] or [15 60 105 150 195 240 285]. The vector of duration should be specified as [15* ones(7,1)] or [15 15 15 15 15 15 15]. - For detrending, select the Wavelet-MDL button. Use the precoloring method: low-pass filter and select the hrf button, and correct for serial correlation, then select the none button.
- In estimating the temporal correlations, check the Individual Analysis.
NOTE: Save the spatial localization of fNIRS channel positions in the individual MNI coordinate systems as a text file. Similarly, save the map based on the individual Brodmann area (BA) as a text file. - In estimating the temporal correlations, check the Group Analysis. The NIRS-SPM aligned the mean optode positions of the number of participants according to the MNI standardized brain coordinate system.
- Compute the activation map based on the changes in hemoglobin level for the standardized brain. HbO2 and HHb levels were considered significant at an uncorrected threshold of p < 0.01.
NOTE: Left/right information was flipped in the right-affected sides for group analysis.
4. Multi-channel comparative analysis based on hierarchical mixed model
- Start the SAS software. Convert the text document (.TXT) of concentration changes of HbO2 and HHb in the NIRS data file processed with a low pass filter (cut-off frequency was set at 0.1 Hz) to the spreadsheet software comma separated values file (.CSV).
- Create the Import SAS data (.sas7bdat) using the program.
- Output the Import file with the following command, libname out "Import file"
- Output the pre- vs. post-intervention file for each subject, run the following commands in the Analysis SAS. (Figure 3) During the creation of the import file, give a name such that can identify the subject information and pre- vs. post-intervention (e.g., id1 pre, id1 post…)
- Run the pre- and post-intervention data command for each channel (ch1-48; HbO2 and HHb) as follows (Figure 4).
- Based on the data obtained from the output results, input the pre- vs. post-intervention differences in change (difference at on-task and rest), rest, and on-task values (estimated values, upper limits, and lower limits) of each channel in the spreadsheet workbook file (.xlsx).
- Similarly, input the numerator and denominator degrees of freedom, F value, and P value of the interact item of the fixed-effect Type 3 test to the spreadsheet workbook file (.xlsx).
- To control the false discovery rate (FDR) in multi-channel testing, use the Benjamin and Hochberg methods27 and control the FDR at a p-value < 0.01.
Representative Results
Herein, we introduce the robot-assisted rehabilitation that our group is currently working on: the biofeedback effects on upper limb motor deficit in patients with acute stroke. We included 10 consenting stroke patients (mean age: 66.8 ± 12.0 years; two women and eight men) who were admitted to our hospital. At the subacute stroke stage, more than 2 weeks after the onset, we evaluated the motor-related cortical activity of these patients using an fNIRS system before and immediately after upper limb robot-assisted rehabilitation on the same day. Regarding the block design tasks, they carried out affected elbow flexion/extension movements 15x within 15 s in each task cycle, and they repeated seven task cycles. In addition, six healthy volunteers (mean age: 58.7 ± 7.1 years; two women and four men) were also enrolled as controls to identify the location of the normal task-related cortical activation during the right elbow flexion/extension movements.
Figure 5 shows the results of the group analysis of 10 stroke patients by the t-statistical mapping values using GLM models with the NIRS-SPM software. This method showed an increase in the cortical activity of the primary motor cortex in the measured hemisphere immediately after the robot-assisted rehabilitation compared with that before the training. The pre-intervention data gathering, intervention (robot-assisted exercise), and the post-intervention data gathering were performed at the single experimental session on the same day and at the same place.
Figure 6 shows the results of the multi-channel group analysis comparing pre- versus post-intervention (robot-assisted exercise). Statistical analysis of the multi-level hierarchical mixed model with the SAS software was performed. Increased cortical activity in the primary motor cortex was observed after intervention, the same brain region as in the NIRS-SPM.
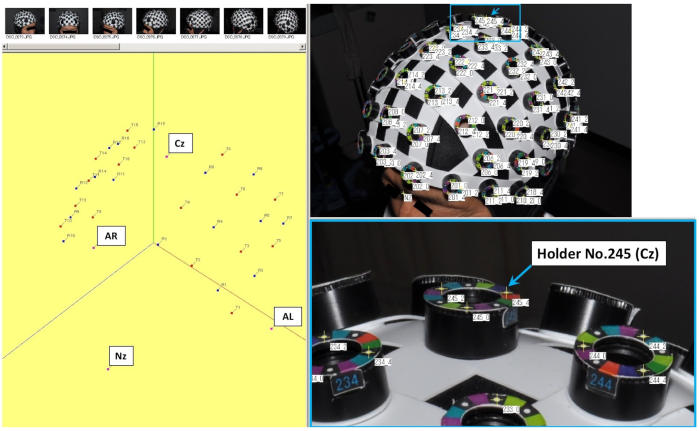
Figure 1: Setting of three-dimensional (3D) spatial registration and each holder of the head cap used for fNIRS recording. The No. 245 holder indicated by the arrow in the figure shows the central position (Cz), which is one of the coordinate axes. Please click here to view a larger version of this figure.
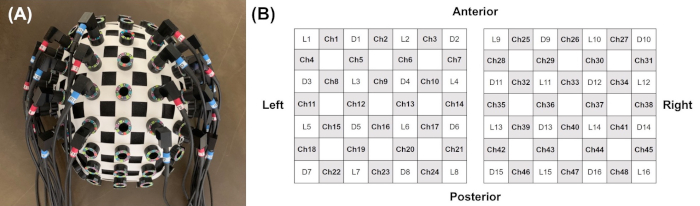
Figure 2: Arrangement of the 48-channel system with 32 optodes during the fNIRS recording. (A) Location of probes on head holder, (B) arrangement of the 48 channels and probes (16 light sources and 16 detectors; 4 x 4 array for each hemisphere) to a head cap bilaterally over the cortical areas, as regions of interest. Please click here to view a larger version of this figure.

Figure 3: Command input for creating files used in the SAS analysis software. The figure shows how to define terms and numerical values on the command input screen that converts the text file information obtained from the NIRS file into an Excel CSV file and then converts it for SAS analysis. ID, Age, Sex, ipsilesional side, before and after the intervention, total time, and task periods, were entered numerically. In addition, information on HbO2, HHb, and total hemoglobin concentration level (mM x mm) were also entered. Please click here to view a larger version of this figure.
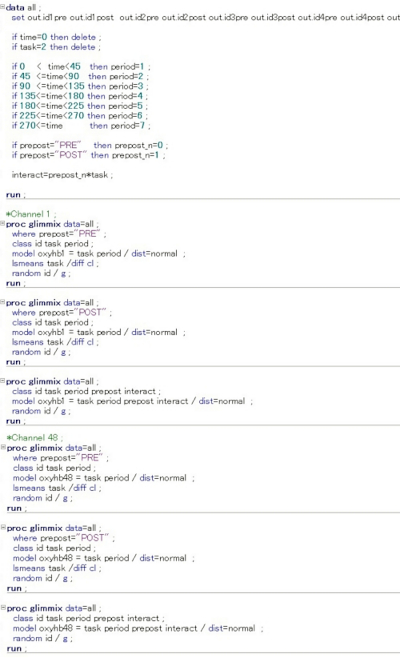
Figure 4: Command input used for each channel analysis in the SAS analysis software. In this multi-level hierarchical mixed model, the following numerical values were set and entered on the SAS command input screen. The status at rest (task = 0) and at task (task = 1) were compared, and the status at recovery (task = 2) was excluded. Furthermore, the status before the intervention was set to n = 0 and the status immediately after the intervention was set to n = 1, and the interaction was investigated for statistically significant differences in the amount of change in HbO2 and HHb for each channel. In the figure, the input screen information up to ch2 or ch47 is omitted. Please click here to view a larger version of this figure.
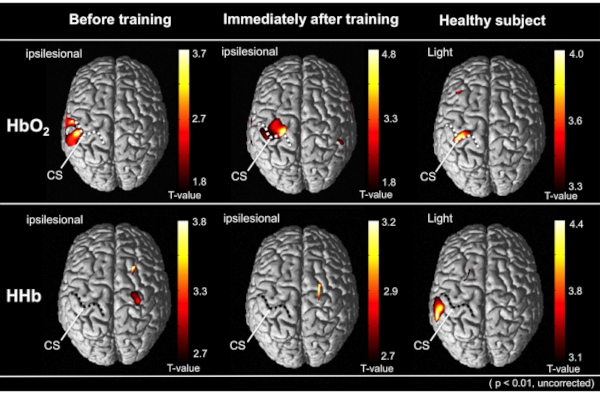
Figure 5: The results of the group analysis by the t-statistical mapping values using GLM models with the NRS-SPM software. The average cortical activity from all patients is depicted on the above view of the standardized brain models. The upper and lower represents the cortical activation in HbO2 and HHb level, respectively. The right image indicates the cortical activity of healthy subjects during the tasks. Comparing the robot-assisted rehabilitation before, the cortical activity was increased immediately after robot-assisted training on the same day. Compared to other cortical regions, each performing status was significantly increased (uncorrected, p < 0.01). Dotted lines indicate the central sulcus (CS) on the normalized brain images. This figure has been modified from Saita et al.15. Please click here to view a larger version of this figure.
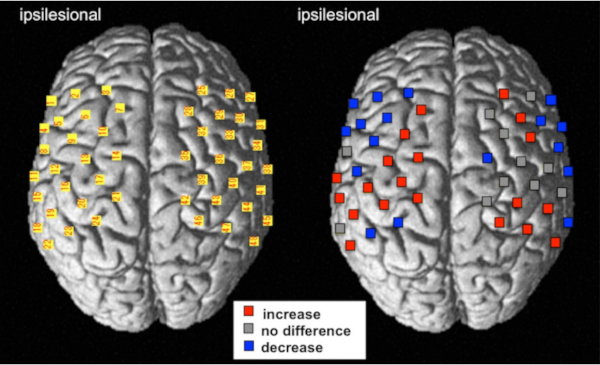
Figure 6: The result of the multi-channel analysis using multi-level hierarchical mixed models with the SAS software. The cortical activity of change represents the comparison between pre-and post-intervention using robot-assisted rehabilitation treatments. For the left image, the numbers of NIRS channels were superimposed on the standardized brain according to the MNI coordinate system. For the right image, red and blue indicate an increase and decrease in the HbO2 level, respectively (FDR corrected, p < 0.01). Gray indicates that the channels did not significantly change following the robot-assisted rehabilitation. This figure has been modified from Saita et al.15. Please click here to view a larger version of this figure.
Discussion
In our group analytic methods for fNIRS, in addition to performing an imaging analytic method by qualitative t-statistic mappings, we compared pre- vs. post-intervention (robot-assisted exercise) using the comparative multichannel analysis. For qualitative analysis, we used the NIRS-SPM software as a mass-univariate approach based on the generalized linear model. The NIRS-SPM analysis shows qualitative results of each session by visualizing the activated area during the task. Moreover, the information of the non-invasive 3D-digitizer enables the estimation of fNIRS channel locations relative to the brain. The group analysis using NIRS-SPM analysis was able to capture the rough brain activational areas of pre- vs. post- intervention during the sensorimotor tasks but could not compare the difference in changes in the same channels. To corroborate the NIRS-SPM findings, the amplitude of the changes in hemoglobin levels induced by the sensorimotor task can be statistically analyzed by comparing the data obtained from two different sessions (i.e., before and after intervention) in the same study subject using the multi-channel hierarchical mixed model. By using these two methods, the results mutually complement each other and were shown more clearly.
To obtain accurate task-related brain activity by NIRS data, task design, probe placement, pre-analysis processing, analysis methods, and environment settings are very important23,26. Regarding the block design using sensorimotor tasks in our representative studies, we set the task and rest time at 15 and 30 s, respectively15. It has been reported that the peak after the activity and the recovery by rest time depend on the task design. In previous researches, it was reported that the task design is often 10-30 s for tasks related to hand movement (finger tapping, grasping task) and 30 s for tasks related to posture control and walking7,8,28. For the task periods, it takes about 5-10 s to reach the peak after starting the task activation8,29, and the recovery periods is favorable with a randomly varying 15 to 18 s to avoid anticipation effects and Mayer-Wave28,30. In this respect, the task protocol of our research is considered to be suitable and feasible as it follows block design with elbow movement. However, the task periods may need to be longer based on the task difficulty, such as for walking tasks and complex cognitive tasks. Regarding the probe arrangement, fNIRS has a poorer spatial resolution, so rearranging pre- vs. post- intervention is a major issue. In our representative study, this shortcoming was compensated by our design not requiring probe relocation to confirm the immediate effect of robotic treatment on the same day. If repositioning is required, it is important to check the distance between the marking sticker and the holder using a pre-captured image to make sure it is not out of arrangement compared to pre-intervention. However, in our design, it was insufficient to confirm the effect of systemic physiological interference such as the autonomic nervous system on the use of the head holder by continuous measurement for a long time. Therefore, it is necessary to use fNIRS monitoring during functional paradigms and multimodal monitoring23 in the future. Regarding the area of interest for NIRS measurement, many NIRS studies on cognitive processing have focused on measuring the prefrontal cortex (PFC) activity given that the PFC is a key area in executive function and cognitive control of movement31,32. For sensorimotor tasks, it is important to measure the parietal region in order to assess sensory activity. The measurement of the parietal region is, however, susceptible to obstacles such as hair and thick scalp; thus, it is necessary to carefully set up the measurement. One limitation of this fNIRS experimental method is that due to the structure of the head holder, we used the general measurement method with an optode distance of 3 cm. However, using short separation channels to calibrate the superficial signals or noises, there is a possibility to measure accurate brain activity33.
Regarding the NIRS group analysis methods, as a prerequisite, it is best to carefully analyze the personal data of NIRS measurement results similar to the EEG. A combination of single-level and group-level analyses may be the optimal approach23. Although the standardized brain is used for the group analysis of NIRS data, limitations with regard to the lower spatial resolution have been discussed4. In this study, the spatial registration method was devised, and it was possible to detect more accurate coordinates, which potentially led to better results. Second, present study has limitations in the ability of the described NIRS system here. The numerical values used in the analysis are relative values using continuous-wave NIRS, and a device such as the Time Domain (TD)-NIRS needs to be used for evaluation using absolute values34,35. However, TD-NIRS is expensive and has a drawback of not being suitable for such multi-channel analysis. Because CW-NIRS is so widely used, we need a relatively accurate evaluation method that can be realized to make up for this shortcoming. As a pre-analysis process, our channel analysis will also need to consider means of using additional principal component analysis to remove these confounders.
In the future, we will report the results of change in pre- vs. post-operation of deep brain stimulation for Parkinson's disease9, cerebrovascular disorders with spasticity12, and cognitive impairment36 using near-infrared spectroscopy application. Our methods can be applied to a variety of neurological disorders such as movement disorders, cerebrovascular diseases, and neuropsychiatric disorders.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was partly supported by the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (C) 18K08956 and a fund from the Central Research Institute of Fukuoka University (No. 201045).
Materials
| 3D-digitizer software | TOPCON | – | NS-1000 software ver.1.50 |
| NIRS system | Shimadzu | – | FOIRE-3000 |
| Robot | CYBERDYNE | – | Single-joint type Hybrid Assitive Limb (HAL-SJ) |
Referências
- Bonilauri, A., Sangiuliano Intra, F., Pugnetti, L., Baselli, G., Baglio, F. A systematic review of cerebral functional near-infrared spectroscopy in chronic neurological diseases-actual applications and future perspectives. Diagnostics (Basel). 10 (8), (2020).
- Mihara, M., Miyai, I. Review of functional near-infrared spectroscopy in neurorehabilitation. Neurophotonics. 3 (3), 031414 (2016).
- Yang, M., Yang, Z., Yuan, T., Feng, W., Wang, P. A systemic review of functional near-infrared spectroscopy for stroke: Current application and future directions. Frontiers in Neurology. 10, 58 (2019).
- Pinti, P., et al. The present and future use of functional near-infrared spectroscopy (fNIRS) for cognitive neuroscience. Annals of the New York Academy of Sciences. 1464 (1), 5-29 (2020).
- Obrig, H. NIRS in clinical neurology – a ‘promising’ tool. Neuroimage. 85, 535-546 (2014).
- Fujimoto, H., et al. Cortical changes underlying balance recovery in patients with hemiplegic stroke. Neuroimage. 85, 547-554 (2014).
- Herold, F., et al. Functional near-infrared spectroscopy in movement science: a systematic review on cortical activity in postural and walking tasks. Neurophotonics. 4 (4), 041403 (2017).
- Leff, D. R., et al. Assessment of the cerebral cortex during motor task behaviours in adults: a systematic review of functional near infrared spectroscopy (fNIRS) studies. Neuroimage. 54 (4), 2922-2936 (2011).
- Morishita, T., et al. Changes in motor-related cortical activity following deep brain stimulation for parkinson’s Disease detected by functional near infrared spectroscopy: A pilot study. Frontiers in Human Neuroscience. 10, 629 (2016).
- Lee, S. H., Jin, S. H., An, J. The difference in cortical activation pattern for complex motor skills: A functional near- infrared spectroscopy study. Science Reports. 9 (1), 14066 (2019).
- Hatakenaka, M., Miyai, I., Mihara, M., Sakoda, S., Kubota, K. Frontal regions involved in learning of motor skill–A functional NIRS study. Neuroimage. 34 (1), 109-116 (2007).
- Saita, K., et al. Combined therapy using botulinum toxin A and single-joint hybrid assistive limb for upper-limb disability due to spastic hemiplegia. Journal of the Neurological Sciences. 373, 182-187 (2017).
- Chang, P. H., et al. The cortical activation pattern by a rehabilitation robotic hand: a functional NIRS study. Frontiers in Human Neuroscience. 8, 49 (2014).
- Bae, S. J., Jang, S. H., Seo, J. P., Chang, P. H. The optimal speed for cortical activation of passive wrist movements performed by a rehabilitation robot: A functional NIRS study. Frontiers in Human Neuroscience. 11, 194 (2017).
- Saita, K., et al. Biofeedback effect of hybrid assistive limb in stroke rehabilitation: A proof of concept study using functional near infrared spectroscopy. PLoS One. 13 (1), 0191361 (2018).
- Mihara, M., et al. Near-infrared spectroscopy-mediated neurofeedback enhances efficacy of motor imagery-based training in poststroke victims: a pilot study. Stroke. 44 (4), 1091-1098 (2013).
- Naseer, N., Hong, K. S. fNIRS-based brain-computer interfaces: a review. Frontiers in Human Neuroscience. 9, 3 (2015).
- Mihara, M., et al. Neurofeedback using real-time near-infrared spectroscopy enhances motor imagery related cortical activation. PLoS One. 7 (3), 32234 (2012).
- Tak, S., Jang, J., Lee, K., Ye, J. C. Quantification of CMRO(2) without hypercapnia using simultaneous near-infrared spectroscopy and fMRI measurements. Physics in Medicine and Biology. 55 (11), 3249-3269 (2010).
- Strangman, G., Culver, J. P., Thompson, J. H., Boas, D. A. A quantitative comparison of simultaneous BOLD fMRI and NIRS recordings during functional brain activation. Neuroimage. 17 (2), 719-731 (2002).
- Scholkmann, F., et al. A review on continuous wave functional near-infrared spectroscopy and imaging instrumentation and methodology. Neuroimage. 85, 6-27 (2014).
- Delpy, D. T., et al. Estimation of optical pathlength through tissue from direct time of flight measurement. Physics in Medicine and Biology. 33 (12), 1433-1442 (1988).
- Tachtsidis, I., Scholkmann, F. False positives and false negatives in functional near-infrared spectroscopy: issues, challenges, and the way forward. Neurophotonics. 3 (3), 031405 (2016).
- Jang, K. E., et al. Wavelet minimum description length detrending for near-infrared spectroscopy. Journal of Biomedical Optics. 14 (3), 034004 (2009).
- Ye, J. C., Tak, S., Jang, K. E., Jung, J., Jang, J. NIRS-SPM: statistical parametric mapping for near-infrared spectroscopy. Neuroimage. 44 (2), 428-447 (2009).
- Orihuela-Espina, F., Leff, D. R., James, D. R., Darzi, A. W., Yang, G. Z. Quality control and assurance in functional near infrared spectroscopy (fNIRS) experimentation. Physics in Medicine and Biology. 55 (13), 3701-3724 (2010).
- Benjamini, Y., Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 57 (1), 289-300 (1995).
- Herold, F., Wiegel, P., Scholkmann, F., Muller, N. G. Applications of functional near-infrared spectroscopy (fNIRS) neuroimaging in exercise(-)cognition science: A systematic, methodology-focused review. Journal of Clinial Medicine. 7 (12), (2018).
- Boden, S., et al. The oxygenation response to functional stimulation: is there a physiological meaning to the lag between parameters. Neuroimage. 36 (1), 100-107 (2007).
- Pinti, P., Scholkmann, F., Hamilton, A., Burgess, P., Tachtsidis, I. Current status and issues regarding pre-processing of fNIRS neuroimaging data: An investigation of diverse signal filtering Methods within a general linear model Framework. Frontiers in Human Neuroscience. 12, 505 (2018).
- Udina, C., et al. Functional near-infrared spectroscopy to study cerebral hemodynamics in older adults during cognitive and motor tasks: a review. Frontiers in Aging Neuroscience. 11, 367 (2019).
- Verghese, J., Wang, C., Ayers, E., Izzetoglu, M., Holtzer, R. Brain activation in high-functioning older adults and falls: Prospective cohort study. Neurology. 88 (2), 191-197 (2017).
- Yucel, M. A., et al. Short separation regression improves statistical significance and better localizes the hemodynamic response obtained by near-infrared spectroscopy for tasks with differing autonomic responses. Neurophotonics. 2 (3), 035005 (2015).
- Torricelli, A., et al. Time domain functional NIRS imaging for human brain mapping. Neuroimage. 85, 28-50 (2014).
- Giacalone, G., et al. Time-domain near-infrared spectroscopy in acute ischemic stroke patients. Neurophotonics. 6 (1), 015003 (2019).
- Saita, K., et al. Contralateral cerebral hypometabolism after cerebellar stroke: a functional near-infrared spectroscopy study. Journal of Stroke and Cerebrovascular Diseases. 26 (4), 69-71 (2017).

