Rapid Freezing using Sandwich Freezing Device for Good Ultrastructural Preservation of Biological Specimens in Electron Microscopy
Summary
Here we show how to use the sandwich freezing device for rapid freezing of biological specimens, including bacteria, yeast, cultured cells, isolated cells, animal and human tissues, and viruses. We also show how to prepare specimens for ultrathin sectioning after rapid freezing.
Abstract
Chemical fixation has been used for observing the ultrastructure of cells and tissues. However, this method does not adequately preserve the ultrastructure of cells; artifacts and extraction of cell contents are usually observed. Rapid freezing is a better alternative for the preservation of cell structure. Sandwich freezing of living yeast or bacteria followed by freeze-substitution has been used for observing the exquisite natural ultrastructure of cells. Recently, sandwich freezing of glutaraldehyde-fixed cultured cells or human tissues has also been used to reveal the ultrastructure of cells and tissues.
These studies have thus far been carried out with a handmade sandwich freezing device, and applications to studies in other laboratories have been limited. A new sandwich freezing device has recently been fabricated and is now commercially available. The present paper shows how to use the sandwich freezing device for rapid freezing of biological specimens, including bacteria, yeast, cultured cells, isolated cells, animal and human tissues, and viruses. Also shown is the preparation of specimens for ultrathin sectioning after rapid freezing and procedures for freeze-substitution, resin embedding, trimming of blocks, cutting of ultrathin sections, recovering of sections, staining, and covering of grids with support films.
Introduction
Electron microscopy is a powerful tool for studying cell ultrastructure. Chemical fixation with conventional dehydration procedures has been used for observing the ultrastructure of cells and tissues. However, this method does not adequately preserve the ultrastructure of cells, and artifacts and the extraction of cell contents are usually observed. Rapid freezing and freeze-substitution of cells and tissues are better alternatives for the preservation of cell structure.
Three main methods have been employed for freezing cells rapidly1: 1) plunge-freezing is performed by plunging specimens into a cooled cryogen, such as propane, and was used since the early 1950s2; 2) cold metal block freezing is performed by rapidly slamming cells and tissues onto a metal block cooled with liquid nitrogen or liquid helium3,4; and 3) high-pressure freezing is done by freezing cells and tissues with liquid nitrogen under high pressure5,6,7.
Sandwich freezing is a type of plunge-freezing carried out by sandwiching thin biological materials between two copper disks and rapidly freezing them by plunging into liquid propane8,9,10. In this method, very thin specimens (a few micrometers thick) are rapidly cooled with cryogen using a metal that has good thermal conductivity from both sides. Thus, this method effectively removes heat from the specimens, making it possible to stably freeze cells without ice-crystal damage. Sandwich freezing, followed by freeze-substitution of living yeast and bacterial cells, reveals the natural ultrastructure of cells10,11,12,13,14,15,16.
Recently, this method has been found to be useful for preserving clear cell images of glutaraldehyde-fixed microorganisms17,18,19,20,21,22,23,24, cultured cells25,26,27, and human cells and tissues1,28. Although these studies have been carried out using a handmade sandwich freezing device29, and applications to other studies in other laboratories have been limited, a new sandwich freezing device (SFD) has been fabricated28 and is now commercially available.
The present paper shows how to use the SFD for rapid freezing of biological specimens, including bacteria, yeast, cultured cells, isolated cells, animal and human tissues, and viruses. Also shown is the preparation of specimens for ultrathin sectioning after rapid freezing, as well as procedures for freeze-substitution, resin embedding, trimming of blocks, cutting of ultrathin sections, recovering of sections, staining, and covering of grids with support films.
Protocol
NOTE: The study's protocol for human samples was approved by the Biomedical Research Ethics Committee of the Graduate School of Medicine, Chiba University (3085). Osmium tetroxide is a dangerous chemical; it should be handled wearing gloves in the fume hood. Figure 1 shows the sandwich freezing device and the necessary tools28. Figure 2 shows the materials necessary to perform sandwich-freezing experiments. Glass vials are filled with acetone containing osmium tetroxide and kept at -80°C until use (Figure 2B). Copper disks are 3 mm in diameter, with no holes, have a letter on one side, and are commercially available (Figure 2C).
1. Rapid freezing of cell suspensions for freeze-substitution
NOTE: The whole procedure is shown in Figure 3.
- Cells
- Use cell suspensions of bacteria, yeast (Figure 2A), cultured cells, and isolated cells for sandwich freezing.
NOTE: Both living and glutaraldehyde-fixed cells can be used28.
- Use cell suspensions of bacteria, yeast (Figure 2A), cultured cells, and isolated cells for sandwich freezing.
- Preparation of liquid propane
NOTE: Use cryogloves and goggles when handling liquid nitrogen. As propane is explosive, care should be taken not to use fires in the same room, and windows should be kept open.- Fill the liquid nitrogen container of the SFD with liquid nitrogen (Figure 4A). Fill the liquid propane container with liquid propane by introducing propane gas using a fine nozzle (Figure 4B,C). Accelerate the solidification of propane by using a cooled metal bar (Figure 1 and Figure 4D,E).
- Preparation of copper disks
- Place copper disks on a slide glass with the no-letter-side up (Figure 2D) and treat with glow discharge at 10 Pa, 400 volts, 1 mA for 30 s (Figure 4F,G) to make the disk surface hydrophilic using an ion sputter apparatus30.
- Sandwiching and plunge-freezing of the cell suspension
- Transfer the cell suspension to a 2-mL centrifuge tube (Figure 5A) and centrifuge at 2,900 × g for 10 s at room temperature (Figure 5B,C). Remove the supernatant, and suspend the pellet to obtain a thick suspension (see discussion).
- Place a small amount of the cell suspension (~0.02 µL) on a copper disk (Figure 5D-F), cover it with another copper disk (Figure 5G), and pick the disks up with tweezers (Figure 5H).
NOTE: To measure ~0.02 µL of the cell suspension, observe 0.1 µL drops of the suspension under the stereomicroscope and divide them into droplets that are 1/5th of this volume. - Make a well in the center of the solid propane with the thin metal bar (Figure 5I). Set the tweezers in an SFD and rapidly freeze them by pushing the injection button of the apparatus (Figure 5J,K).
NOTE: Care should be taken not to dry the specimens and not lay the two disks completely over each other (otherwise, detaching them would become very difficult in the next step).
- Sandwiching and plunge-freezing of animal and human tissues
- Use animal and human tissues (~0.5 mm x 0.5 mm x 1.5 mm) fixed in 2.5% glutaraldehyde-0.1 M phosphate buffer (pH 7.4) (Figure 6A,B). Slice them into 0.1- to 0.2- mm thick sections with a razor blade under a stereomicroscope (Figure 6C,D).
- Place a small drop (~0.02 µL) of glutaraldehyde solution on a copper disk (Figure 6E, F). Then, use tweezers to place a piece of tissue in the glutaraldehyde on the copper disk (Figure 6G,H) and cover it with another copper disk (Figure 6I-K).
- Rapidly freeze the disks with the tissues in the melting propane of the SFD, as described in section 1.4 (Figure 5J,K).
NOTE: As glutaraldehyde is a dangerous chemical, it should be handled wearing gloves in the fume hood. Tissues should not be washed with buffers when placed on a copper disk but kept soaked in glutaraldehyde solution because glutaraldehyde has an anti-freezing effect1.
- Use animal and human tissues (~0.5 mm x 0.5 mm x 1.5 mm) fixed in 2.5% glutaraldehyde-0.1 M phosphate buffer (pH 7.4) (Figure 6A,B). Slice them into 0.1- to 0.2- mm thick sections with a razor blade under a stereomicroscope (Figure 6C,D).
- Freeze-substitution with osmium acetone
- Transfer the disks into liquid nitrogen in a working bath (Figure 7A,B). Using a pair of tweezers cooled in liquid nitrogen, detach the disks from each other to expose the specimen (Figure 7C-E).
- Place the disks with the cells in a glass vial (Figure 7F) that is filled with 1 mL of acetone containing 2% osmium tetroxide (Figure 2B) that has been placed in liquid nitrogen and solidified (Figure 4H).
- Transfer the disks to a deep-freezer and keep them at -80°C for 2-4 days for freeze-substitution of the cells (Figure 3). Soak the used tweezers in water at room temperature to warm them (Figure 7G) for the freezing of the following specimens.
NOTE: Tweezers for handling the specimen should be warm (room temperature) because cold tweezers may freeze the specimen before rapid freezing, leading to the formation of ice crystals.
- Sample warming and embedding
- Gradually bring the specimens to room temperature (2 h at -20°C, 2 h at 4°C, and 15 min at room temperature, Figure 3 and Figure 8A), and transfer the disks to 2-mL plastic tubes that are filled with 1 mL of acetone (Figure 3 and Figure 8B,C).
- Prepare epoxy resin by mixing the reagents in a disposable plastic container using a stirrer (Figure 8D–F).
- Exchange the acetone in step 1.7.1 successively with 30% resin (in acetone), 60% resin, and 90% resin at room temperature for 1 h each. Then, exchange the 90% resin with 100% resin at 37°C overnight. Finally, embed the samples in 100% resin (Figure 8G-J) in the silicon embedding mold, and polymerize them at 60°C for 24 h (Figure 3).
NOTE: The specimens should remain attached to the copper disks throughout the entire procedure (to make sectioning easier). The use of a rotating or shaking apparatus is not recommended during the embedding process because they contribute little to the penetration of the resin into the cell. Moreover, the vibration from the apparatus sometimes causes the specimens to detach from the copper disks. Incubation at 37°C overnight would accelerate the penetration of the resin into the cell due to thermal energy (the resin would not polymerize because it does not contain any accelerator).
- Trimming of specimen blocks
- Take out the polymerized blocks from the silicon embedding molds (Figure 9A). Write the specimen number on the block (Figure 9B).
- Remove the copper disks from the block with a razor blade (Figure 9C,D) and trim the specimens embedded on the block surface to 0.7 mm x 0.7 mm by using an ultrasonic trimming blade (Figure 9E) and razor blades (Figure 9F-H) under a stereomicroscope31.
- Set the block in a specimen holder of an ultramicrotome (Figure 9I) and cut the face of the block smoothly with a diamond trimming knife (Figure 9J,K).
- Cutting ultrathin sections
- Remove the block from the ultramicrotome (Figure 10A)31, set it in a stereomicroscope, and trim it further to 0.2 mm x 0.3 mm with a razor blade (Figure 10B,C).
- Apply neoprene on the grids to make them adhesive (Figure 10D). Set the block back on the ultramicrotome, cover the ultramicrotome with a plastic cover (Figure 10E)31, and cut 50-70-nm-thick sections (Figure 10F-H).
- Retrieve the sections using a loop (Figure 10I), mount them on 300 or 400 mesh copper grids treated with neoprene, and dry them (Figure 10J).
- Staining of sections and observation under the electron microscope
- Set the grids with sections in the groove of the half silicone tube (Figure 11A)31, and soak in uranyl acetate solution and lead citrate for 3 min each for staining (Figure 11B-E)32.
- For yeast and fungal specimens, place the grids on a filter paper (4 mm x 4 mm), pick them up with tweezers, and cover them with plasma-polymerized naphthalene film33 on the surface of the water (Figure 11F,G). Insert the grids into an electron microscope and observe at 100 kV (Figure 11H,I).
2. Rapid freezing of viruses and macromolecules
- Preparation of liquid ethane
NOTE: Use cryogloves and goggles when handling liquid nitrogen. As ethane is explosive, care should be taken not to use fires in the same room, and windows should be kept open. Ethane is used because it evaporates in the electron microscope while propane does not.- Fill the liquid nitrogen container of the SFD with liquid nitrogen. Fill the liquid ethane container with liquid ethane by introducing ethane gas through a fine nozzle.
- Preparation and rapid freezing of microgrids and specimens
- Make both faces of the microgrids hydrophilic by treating them with glow discharge (10 Pa, 400 V, 1 mA) using an ion sputter apparatus30.
- Set the microgrid in the SFD and apply 2 µL of the virus or macromolecular suspension (1 mg protein/mL) on the microgrids. Remove excess liquid using filter paper and rapidly freeze the microgrid by pushing the injection button of the apparatus.
- Setting of the frozen microgrid in a cryo-transfer holder and observation under the electron microscope
- Transfer the frozen microgrids in liquid nitrogen, set in a cryo-transfer holder cooled at liquid nitrogen temperature beforehand, and observe under an electron microscope at low temperature34.
Representative Results
Living cells of microorganisms in suspension were collected by centrifugation, sandwiched between two copper disks, rapidly frozen with SFD, freeze-substituted, embedded in epoxy resin, ultrathin-sectioned, stained, and observed under an electron microscope by following the procedures described above. Figure 12 shows ultrathin sections of Escherichia coli (bacteria, Figure 12A,B)16 and Saccharomyces cerevisiae (yeast, Figure 12C)15. Note that the images are very clear and show natural morphology.
Glutaraldehyde-fixed cell suspensions of cultured cells and isolated animal cells were collected and rapidly frozen with SFD, freeze-substituted, and observed under an electron microscope by following the procedures described above. Figure 13 shows ultrathin sections of cultured cells (Figure 13A–C)1,28 and isolated cells from mouse abdominal cavity (Figure 13D)28. Figure 14 shows ultrathin sections of human skin (Figure 14A–D) and buffy coat (Figure 14E)1. Note that the images are also very clear and show natural morphology. Figure 15 shows hepatitis B virus core particles rapidly frozen with SFD and observed by cryo-electron microscopy34. As with the other cells, the images are very clear and show natural morphology.
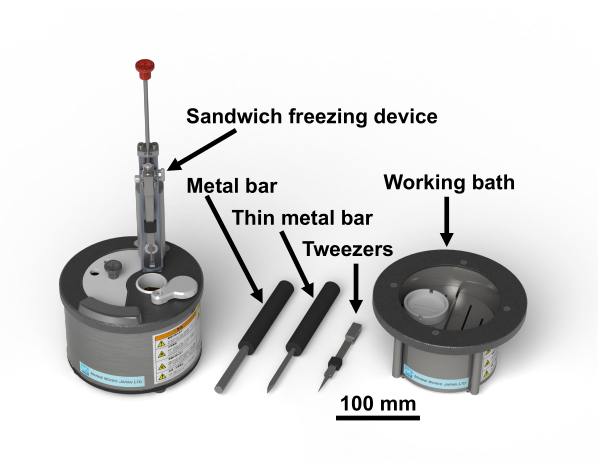
Figure 1: Sandwich freezing device28 and the necessary tools. Please click here to view a larger version of this figure.
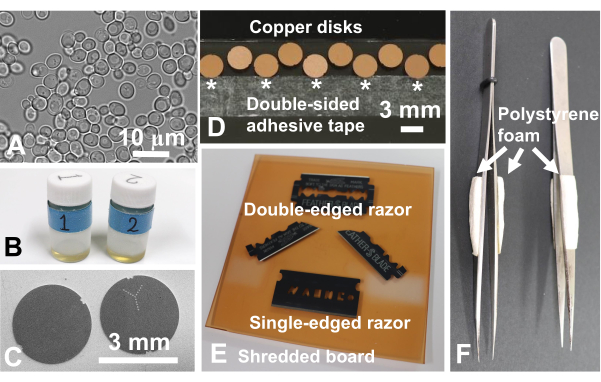
Figure 2: Materials necessary to perform sandwich freezing experiments. (A) Specimen: Light micrograph of Saccharomyces cerevisiae (yeast). Scale bar = 10 µm. (B) Glass vials (10 mL) containing 1 mL of acetone with 2% osmium tetroxide. (C) Copper disks showing a surface without a letter (left) and a surface with a letter (right). Scale bar = 3 mm. Scanning electron microscopy. (D) Copper disks with no-letter-side up were placed on a glass slide with double-sided adhesive tape (*). Scale bar = 3 mm. (E) A double-edged razor and a broken double-edged razor for slicing animal and human tissues, a single-edged razor for trimming blocks, and shredded board for slicing animal and human tissues. (F) Tweezers with extruded polystyrene foam to protect fingers from the cold. Please click here to view a larger version of this figure.
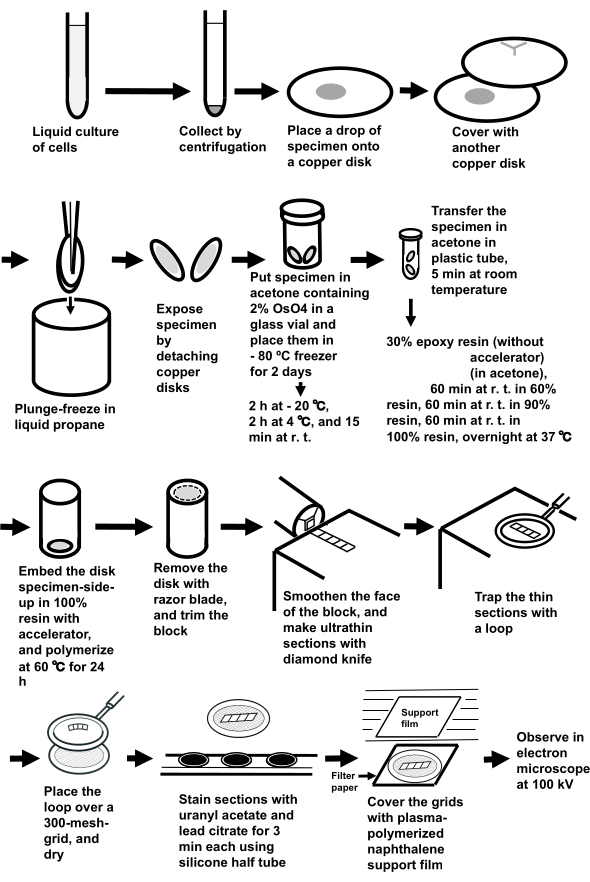
Figure 3: Specimen preparation of cell suspensions using the sandwich freezing method. Abbreviations: r.t. = room temperature. Please click here to view a larger version of this figure.

Figure 4: Preparation of liquid propane, copper disks, and fixative. (A) Liquid nitrogen (arrow) was poured into the liquid nitrogen container of the sandwich freezing device. (B) Propane gas was introduced into a liquid propane container through a fine nozzle. (C) Liquid propane (arrow). (D) A metal bar (arrow) was used to cool liquid propane to accelerate the solidification of liquid propane. (E) Solidified propane (arrow). (F) Ion sputter apparatus (arrow) for making copper disks hydrophilic with glow discharge. (G) Glow discharge. (H) Glass vials placed in liquid nitrogen in a working bath. Please click here to view a larger version of this figure.

Figure 5: Rapid freezing of cell suspension (yeast) using the sandwich freezing device. (A) Transfer of yeast culture into the centrifuge tube. (B) Microcentrifuge. (C) Pellet of Saccharomyces cerevisiae in the centrifuge tube (arrow). (D) Transferring the specimen with a micropipette from the centrifuge tube. (E) Placing the specimen on the copper disk. (F) A small drop of a specimen on the copper disk (arrow). (G) Covering the specimen with another copper disk. (H) Picking the two disks up with tweezers. (I) Making a well in the solid propane using the thin metal bar. (J) Plunge-freezing of the specimen by pushing the injection button. (K) Freezing is complete. Please click here to view a larger version of this figure.
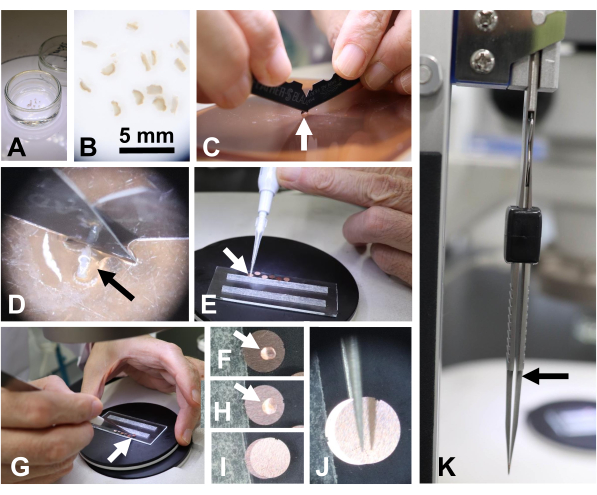
Figure 6: Preparing a specimen of human tissues (skin). (A, B) Human skin tissue fixed in glutaraldehyde in a Petri dish. Scale bar = 5 mm. (C, D) Tissues (arrow) were sliced using two double-edged razors on a shredded board. (E, F) A small drop of glutaraldehyde solution (arrow) was placed on a copper disk. (G, H) A piece of skin tissue was placed on the copper disk using tweezers. (I) Another disk was used to cover the copper disk with the skin tissue. (J) The sandwiched disks were picked up with tweezers. (K) The sandwiched disks were gently held with tweezers. Note the gap between the tweezer tips (arrow) maintained to avoid crushing the tissues. Please click here to view a larger version of this figure.
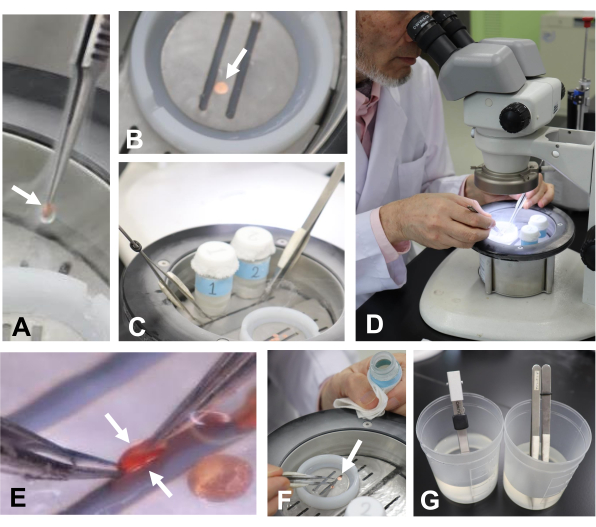
Figure 7: Transferring the specimen into liquid nitrogen and detaching the disks. (A) Transferring the disks (arrow) into liquid nitrogen in a working bath. (B) Disks in liquid nitrogen (arrow). (C) Tweezers were placed in liquid nitrogen to cool them in the working bath. (D, E) Detaching the copper disks (arrows) using tweezers. (F) Transferring the disk (arrow) into the glass vial with tweezers. (G) Warming the tweezers in water for freezing the next specimen. Please click here to view a larger version of this figure.

Figure 8: Sample warming and embedding. (A) Specimens in glass vials at room temperature. (B) Transferring the disks (arrow) into a 2 mL plastic tube with tweezers. (C) Copper disks (arrow) in acetone in a plastic tube. (D) Measuring resins with an injection tube (arrow). (E) Transferring the resin into a disposable cup (arrow). (F) Mixing the resin using a stirrer. (G) A small amount of resin was placed in the holes of the silicone embedding mold. (H) Excess resin in the grids was removed with filter paper. (I, J) Copper disks with specimens were placed in the bottom of the holes of the embedding mold with the specimen side up. Please click here to view a larger version of this figure.
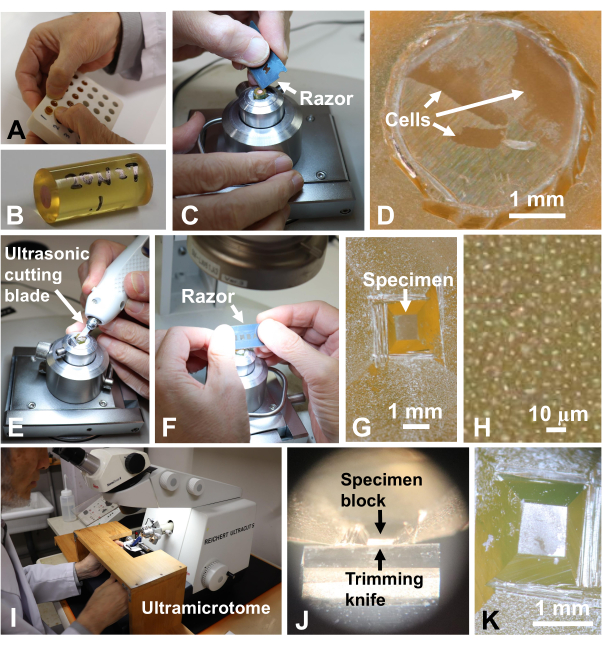
Figure 9: Trimming the specimen block. (A) Taking out polymerized blocks from the embedding molds. (B) Specimen number was written on the block. (C, D) The copper disk was removed from the block with a razor. Scale bar for (D) = 1 mm. (E–G) The specimen was trimmed with an ultrasonic cutting blade and a razor blade. Scale bar for (G) = 1 mm. (H) High magnification of (G). An individual bright spot is a cell. Cells were embedded in a single layer10. Scale bar = 10 µm. (I–K) The surface of the block was cut smooth with a diamond trimming knife. Scale bar for (K) = 1 mm. Please click here to view a larger version of this figure.
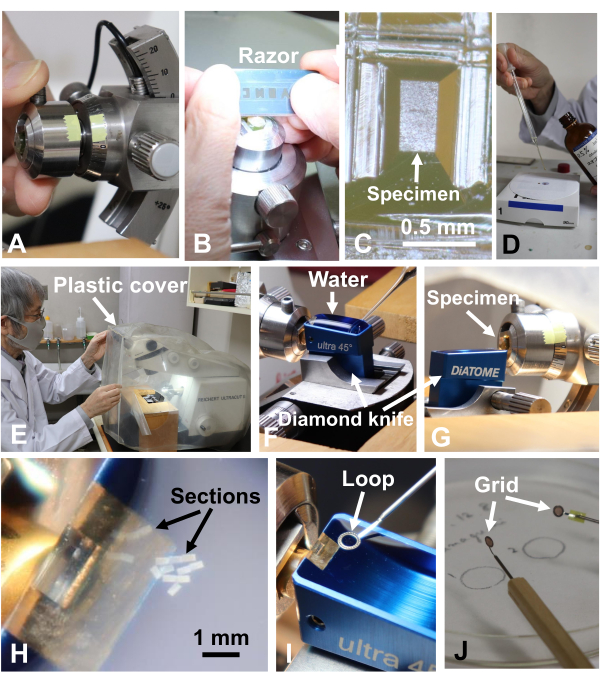
Figure 10: Cutting ultrathin sections. (A) The specimen block was removed from the microtome31. (B, C) The specimen was further trimmed with a razor blade. Scale bar for (C) = 0.5 mm. (D) Neoprene was applied on grids to make them stick. (E) The ultramicrotome was covered with plastic to avoid airflow during ultrathin sectioning. (F) The diamond knife boat was filled with water. (G, H) Ultrathin sections were cut to a thickness of 70 nm. Scale bar for (H) = 1 mm. (I, J) The sections were retrieved using a loop and then dried. Please click here to view a larger version of this figure.
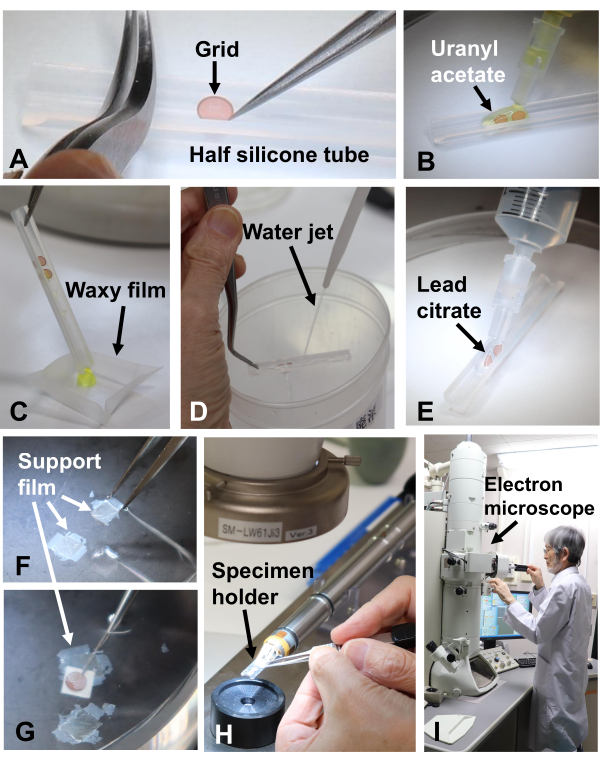
Figure 11: Staining sections. (A) The grids were placed in the groove of the half silicone tube. (B) They were soaked in uranyl acetate for staining. (C) The used uranyl acetate was collected on self-sealing waxy film. (D) The grids were washed with a water jet and (E) then soaked in lead citrate. (F) Plasma-polymerized naphthalene film was floated on water and (G) used to cover a grid placed on 4 mm x 4 mm filter paper. (H) The grid was placed in a specimen holder and (I) observed in an electron microscope. Please click here to view a larger version of this figure.
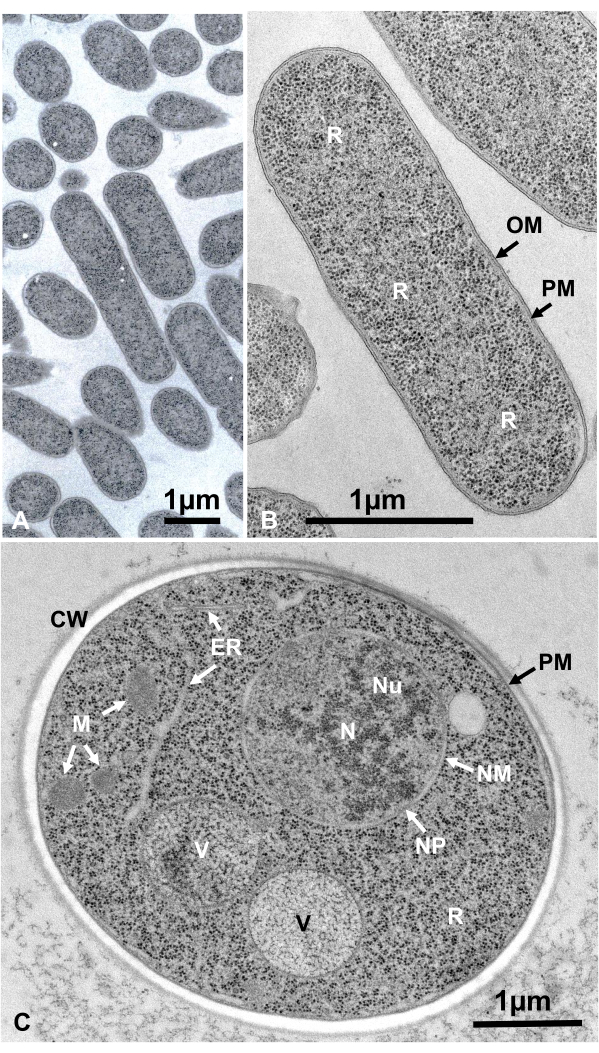
Figure 12: Ultrathin sections of Escherichia coli (bacteria, A, B) and Saccharomyces cerevisiae (yeast, C). (A) Note that specimens are embedded densely and homogeneously and show no deformation. (B, C) Membrane structures show clear and smooth morphology, and ribosomes are clear enough that every particle can be enumerated16. (C) Yeast nucleus and vacuoles show a true circle shape, which may be their natural morphology. The matrix of the mitochondria shows an electron-dense appearance, which may be a characteristic of live cells that are fixed by rapid freezing. Scale bars = 1 µm. Abbreviations: CW = cell wall; ER = endoplasmic reticulum; NM = nuclear membrane; NP = nuclear pores; OM = outer membrane; PM = plasma membranes; R = ribosomes; N = nucleus; M = mitochondria. (B) is reproduced from Yamada et al.16 with permission. Please click here to view a larger version of this figure.
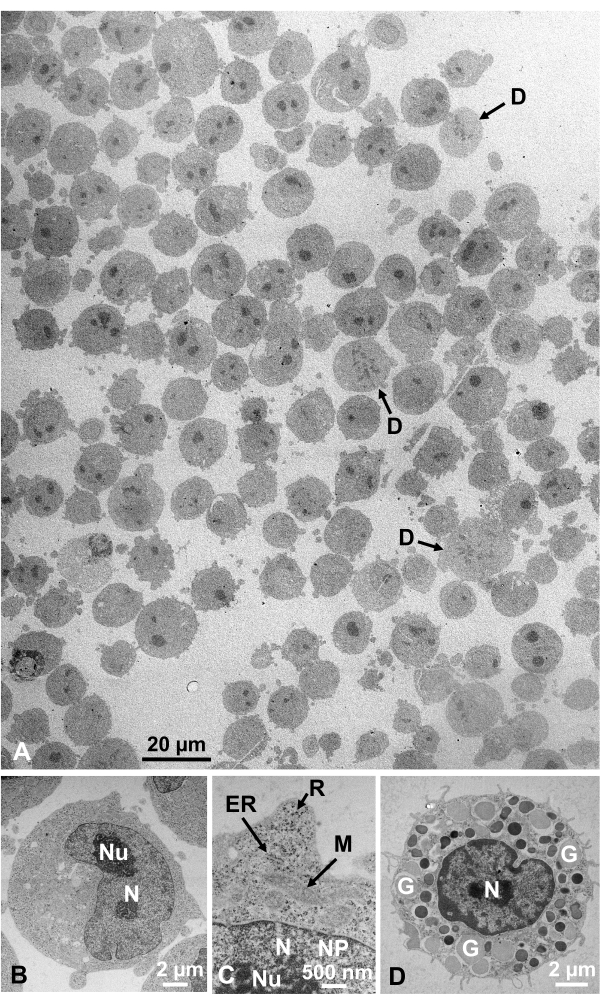
Figure 13: Ultrathin sections. (A–C) K562 cultured cells. (D) An isolated mast cell from mouse. (A) Note that specimens are embedded densely and homogeneously and show no deformation. Scale bar = 20 µm. (B–D) At high magnification, nucleus, nucleolus, nuclear membrane, nuclear pores, endoplasmic reticulum, mitochondria, ribosomes, and granules are clearly observed. Scale bars = 2 µm (B), 500 nm (C), 2 µm (D). Abbreviations: N = nucleus; Nu = nucleolus; NM = nuclear membrane, NP = nuclear pores; ER = endoplasmic reticulum; M = mitochondria; R = ribosomes; G = granules; D = dividing cells. (A) is reproduced from Yamaguchi et al.1 with permission. (B–D) are reproduced from Yamaguchi et al.28 with permission. Please click here to view a larger version of this figure.
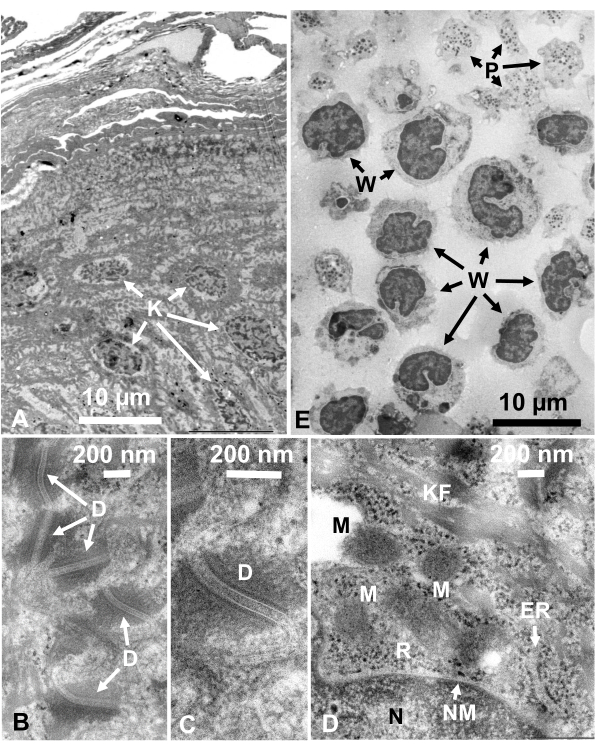
Figure 14: Ultrathin sections. (A–D) human skin and (E) buffy coat. (A–E) Note that tissue and cell images are clear and natural and show good contrast, although the sections are very thin (50 nm). The matrix of the mitochondria shows a dense appearance, which is similar to those of dense mitochondrial matrices of rapidly frozen living cells (D). Scale bars = 10 µm (A, E), 200 nm (B–D). Abbreviations: D = desmosomes; ER = endoplasmic reticulum; K = keratinocyte; KF = keratin fibers; M = mitochondria; N = nucleus; NM = nuclear membrane; P = platelet; R = ribosomes; W = white blood cells. Reproduced from Yamaguchi et al.28 with permission. Please click here to view a larger version of this figure.
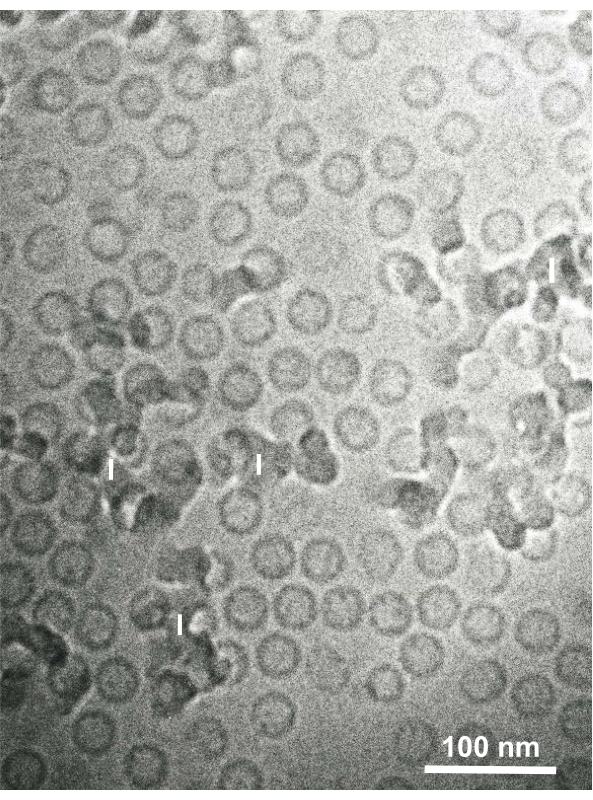
Figure 15: Hepatitis B virus (HBV) core particles rapidly frozen with liquid ethane using the sandwich freezing device and observed by cryo-electron microscopy. Spherical hollow particles are HBV core particles. Scale bar = 100 nm. Abbreviations: HBV = hepatitis B virus; I = ice. Reproduced from Yamaguchi et al.28 with permission. Please click here to view a larger version of this figure.
Discussion
The following discussion is based on more than 120 sandwich freezing-freeze substitution experiments on more than 1,000 samples and more than 70 plunge-freezing-cryo-electron microscopy experiments on more than 75 samples conducted over 36 years.
Success rate for good freezing by sandwich freezing
The rate of success in achieving good freezing depends on the samples. Saccharomyces cerevisiae (yeast) cells cultured in YPD medium (1% yeast extract, 2% peptone, 2% dextrose) gave nearly 100% success for good freezing without ice crystal formation10,11,15,35,36. Other yeast species, including Shizosaccharomyces37,38,39, Cryptococcus14,40,41,42,43,44,45, Exophiala13,41,46,47,48,49, Fusarium50,51,52, Aureobasidium53, Candida54,55, Fellomyces56, Aspergillus57, and Trichosporon, also showed good freezing. Bacteria, including Mycobacterium58,59 and E. coli16, also showed good freezing. Cultured cells and isolated animal cells showed good freezing for both living and glutaraldehyde-fixed cells1,25,26,27,60. Glutaraldehyde-fixed animal and human tissues sliced to 0.1 to 0.2 mm thickness also showed good freezing most of the time1,28.
Conditions for good freezing
Use only cells in the appropriate growth stage and condition. Cells in culture should be in the exponential phase. Apply very small amounts of cell suspensions of concentrated samples (for S. cerevisiae, ~0.02 µL of 3-5 × 109 cells/mL) on the copper disk. Glutaraldehyde-fixed slices of animal or human tissues should also be very small (preferably 0.3 mm x 0.3 mm x 0.1 mm). Because cutting 0.1-mm thick tissue slices is difficult, slice many tissues and select thin and half-transparent slices. Work quickly but carefully, and do not let the samples dry out. In picking up the stacked copper disks with tweezers, do not press the disks too hard to avoid crushing the cells and tissues. Specimen loading is the most important step for successful freezing, and the conditions required are the same as the conditions for good freezing for high-pressure freezing. Readers should refer to the excellent review by McDonald61.
Other applications
This paper presents electron micrographs of a bacterium, yeast, cultured cells, isolated animal cells, human tissues, and virus particles. We observed good freezing of glutaraldehyde-fixed marine algae. However, the cell structures of living freshwater green algae were destroyed by ice crystal formation. Mixing cells with 20% bovine serum albumin (BSA) ensured that the ultrastructure was well-preserved with no ice-crystal damage. The use of 20% BSA was also beneficial for preserving the ultrastructure of stalk cells of a mushroom. Experiments on the freezing of plant cells and tissues by applying 20% BSA are ongoing. Although scanning electron microscopy of sandwich freezing-freeze-substituted samples has not been attempted, observation of well-preserved cell structures has been reported previously9.
Notes on the sandwich freezing method
The close-to-native ultrastructure of cells is best observed by rapid freezing and freeze-substitution of living cells. Ice crystal formation with the SFD can be avoided by limiting the thickness of cells to ≤30 µm1. Fixing tissues with glutaraldehyde often yields better preservation of the cell structure for observing suspension-cultured cells because glutaraldehyde fixation makes the cell structure more rigid and prevents the possible ultrastructural changes during collection and centrifugation of living cells1. Glutaraldehyde fixation also allows the extension of the freezing depth to as much as 0.2 mm1, similar to that achieved by the high-pressure freezing (HPF) method. Therefore, the HPF machine can be replaced with the SFD for deep freezing of animal and human tissues.
Because glutaraldehyde-fixed tissues can be stored for more than 2 years28, sandwich freezing can be performed according to the user's convenience. Fixing tissues with glutaraldehyde also facilitates tissue sectioning because the tissues become more rigid with fixation. Unlike the HPF machine, the SFD can be used for rapid freezing of viruses for cryo-electron microscopy and for bacteria and eukaryotic cells. Moreover, compared to the HPF machine, the SFD is small, portable, less expensive, and can be acquired by more laboratories. We hope that these features of the SFD help more laboratories achieve their research goals28.
Features of the natural morphology of cells
Cell structures are in their natural state if they show the following appearance: Membrane structures of the outer membrane (Figure 12A, B), plasma membrane (Figure 12B,C; Figure 13A–D; and Figure 14E), nuclear envelope (Figure 12C, Figure 13B–D, and Figure 14D–E), mitochondria (Figure 12C, Figure 13C, and Figure 14D), and vacuoles (Figure 12C) show smooth contours. The nucleus and vacuoles are almost circular (Figure 12C). Ribosomes show a clear electron-dense appearance with a diameter of ~20 nm (Figure 12B,C; Figure 13C; and Figure 14D). The cytoplasm is electron-lucent (Figure 12B,C; Figure 13C; and Figure 14D).
Effect of glutaraldehyde fixation on cell morphology
Glutaraldehyde fixation was performed for animal or human tissues before sandwich freezing to obtain ice-crystal-free ultrastructure. The micrographs obtained by this method show exquisitely clear images similar to the ones obtained by the rapid freezing of living tissues (Figure 14)1. The studies on yeast cells, deep-sea microorganisms, and cultured cells show that the deformation of ultrastructure is mainly due to osmium tetroxide fixation and dehydration by ethanol at room temperature1,17,18,28. Small also reported that although glutaraldehyde fixation does not destroy the organization of actin in cultured fibroblasts, osmium tetroxide fixation and dehydration by acetone or ethanol at room temperature destroy actin organization62.
Hence, a detailed study on the effects of glutaraldehyde fixation on cell morphology should be carried out. Ohno developed an in vivo cryofixation method wherein living tissues are rapidly frozen without stopping the blood supply63. The tissues were freeze-substituted and embedded in epoxy resin, and ultrathin sections were observed. The electron microscopic images showed the closest-to-native ultrastructure of living tissues compared with those obtained by chemical fixation-conventional dehydration and by the rapid freezing of fresh unfixed tissues. Therefore, it may be interesting to compare the ultrastructure obtained by glutaraldehyde fixation-freeze substitution (the present method) and those by in vivo cryofixation-freeze substitution to examine the effects of glutaraldehyde fixation.
Consideration for the environment and increased experimental efficiency
We use 2 mL plastic tubes for the substitution of the resin. One mL of diluted resin is enough for each substitution step. The used plastic tubes may be discarded after each experiment. This can save time and effort for washing glass vials when they are used for resin substitution. Additionally, uranyl acetate solution can be used repeatedly for section staining32. After staining the sections, the uranyl acetate solution can be saved and re-used. As uranyl acetate is a radioactive substance, its re-use helps avoid the generation of waste and contributes to the protection of the environment.
Plasma-polymerized naphthalene film
Plasma-polymerized naphthalene film is a three-dimensionally polymerized carbon film made from naphthalene gas by plasma-polymerization under glow discharge33. The film is resilient against electron bombardment and chemicals, very clean, transparent against electrons, and has a flat surface and amorphous structure. Thus, the plasma-polymerized naphthalene film, which is commercially available, is excellent and is recommended as a support film.
Declarações
The authors have nothing to disclose.
Acknowledgements
None
Materials
| Sandwich Freezing Device | Marine works Japan, Ltd, Yokosuka, Japan | MW-SFD-01 | with metal bar, thin metal bar, tweezers, and working bath |
| 10 mL glass vials | – | Scintillation counter vials for fixative | |
| Acetone | – | ||
| Osmium tetroxide | Nisshin EM Co. Ltd., Tokyo | 3004 | 0.1 g |
| Deep freezer | Sanyo Co. Ltd., Osaka | MDF-C8V1 | |
| Copper disk | Nisshin EM Co. Ltd., Tokyo | – | Refer to this paper |
| Slide glass | – | ||
| Double-sided adhesive tape | – | ||
| Single-edged razor blade | Nisshin EM Co. Ltd., Tokyo | – | Feather, FAS-10 |
| Double-edged razor blade | Nisshin EM Co. Ltd., Tokyo | – | Feather, FA-10 |
| Shredded board | Nisshin EM Co. Ltd., Tokyo | 428 | |
| Tweezers | Nisshin EM Co. Ltd., Tokyo | – | Several pairs |
| Tweezers with polystyrene foam | – | One pair | |
| Glutaraldehyde | Nisshin EM Co. Ltd., Tokyo | 3052 | |
| Liquid nitrogen | – | ||
| Propane gas | – | Cryogen | |
| Ion sputter apparatus | Hitachi high technologies, Tokyo | Hitachi E102 | |
| Micropipette | – | For 1 mL, 200 μL, and 2 μL | |
| Microcentrifuge | Tomy digital biology Co. Ltd., Tokyo | Capsulefuge, PMC-060 | |
| Stereomicroscope | Nikon Co. Ltd., Tokyo et al. | – | SMZ 645 |
| LED illumination for stereomicroscope | Nikon Co. Ltd., Tokyo et al. | SM-LW 61 Ji | |
| Disposable plastic container | – | 50 mL and 200 mL | |
| Ethane gas | – | Cryogen | |
| 2 mL Eppendorf tubes | – | For embedding | |
| Disposable plastic syringes | – | 1 mL, 5 mL, 10 mL, and 20 mL | |
| Magnetic stirrer | – | ||
| Epoxy resin | Nisshin EM Co. Ltd., Tokyo | 340 | Quetol 812 set |
| Silicon embedding mold | Nisshin EM Co. Ltd., Tokyo | 4217 | 7 mm in diameter, 13 mm deep |
| Incubater | – | For 37 °C and 60 °C | |
| Trimming stage | Sunmag Co. Ltd., Tokyo | – | Tilting mechanism equipped, Refer to this paper |
| LED illumination for trimming stage | Sunmag Co. Ltd., Tokyo | – | Refer to this paper |
| Ultrasonic Trimming Blade | Nisshin EM Co. Ltd., Tokyo | 5240 | EM-240, Refer to this paper |
| Ultramicrotome | Leica Microsystems, Vienna | Ultracut S | |
| Grids | Nisshin EM Co. Ltd., Tokyo | 2633, 2634 | 300 mesh, 400 mesh |
| 0.5% Neoprene W solution | Nisshin EM Co. Ltd., Tokyo | 605 | |
| Perfect Loop | Nisshin EM Co. Ltd., Tokyo | 2351 | Fot retrieving sections |
| Half Tube for section staining | Nisshin EM Co. Ltd., Tokyo | 463 | Refer to this pape |
| Syringe filter | Toyo Roshi Kaisha, Ltd., Tokyo | DISMIC-03CP | Cellulose acetate, 0.45 μm |
| Transmission electron microscope | JEOL Co. Ltd., Tokyo | JEM-1400 |
Referências
- Yamaguchi, M., et al. Good ultrastructural preservation of human tissues and cultured cells by glutaraldehyde fixation, sandwich freezing, and freeze-substitution. Cytologia. 85 (1), 15-26 (2020).
- Gilkey, J. C., Staehelin, L. A. Advances in ultrarapid freezing for the preservation of cellular ultrastructure. Journal of Electron Microscopy Technique. 3 (2), 177-210 (1986).
- Van Harreveld, A., Crowell, J. Electron microscopy after rapid freezing on a metal surface and substitution fixation. The Anatomical Record. 149, 381-385 (1964).
- Aoki, N., Ito, M., Ejiri, S., Ozawa, H. Ultrastructure of human skin by a rapid freezing technique: structural preservation and antigenicity. Journal of Investigative Dermatology. 102 (3), 354-361 (1994).
- Moor, H., Steinbrecht, R. A., Zierold, K. Theory and practice of high pressure freezing. Cryotechniques in Biological Electron Microscopy. , 175-191 (1987).
- McDonald, K. L., Morphew, M., Vertkade, P., Muller-Reichert, T. Recent advances in high-pressure freezing: Equipment-and specimen-loading methods. Methods in Molecular Biology. 369, 143-173 (2007).
- Sosinsky, G. E., et al. The combination of chemical fixation procedures with high pressure freezing and freeze substitution preserves highly labile tissue ultrastructure for electron tomography applications. Journal of Structural Biology. 161 (3), 359-371 (2008).
- Costello, M. J. Ultra-rapid freezing of the biological samples. Scanning Electron Microscopy. , 361-370 (1980).
- Baba, M., Osumi, M. Transmission and scanning electron microscopic examination of intracellular organelles in freeze-substituted Kloeckera and Saccharomyces cerevisiae yeast cells. Journal of Electron Microscopy Technique. 5 (3), 249-261 (1987).
- Yamaguchi, M., Okada, H., Namiki, Y. Smart specimen preparation for freeze-substitution and serial ultrathin sectioning of yeast cells. Journal of Electron Microscopy. 58 (4), 261-266 (2009).
- Yamaguchi, M., et al. Electron microscopy of hepatitis B virus core antigen expressing yeast cells by freeze-substitution fixation. European Journal of Cell Biology. 47 (1), 138-143 (1988).
- Baba, M., Takeshige, K., Baba, N., Ohsumi, Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. Journal of Cell Biology. 124 (6), 903-913 (1994).
- Yamaguchi, M., et al. The spindle pole body duplicates in early G1 phase in a pathogenic yeast Exophiala dermatitidis: an ultrastructural study. Experimental Cell Research. 279 (1), 71-79 (2002).
- Yamaguchi, M., Biswas, S. K., Ohkusu, M., Takeo, K. Dynamics of the spindle pole body of the pathogenic yeast Cryptococcus neoformans examined by freeze-substitution electron microscopy. FEMS Microbiology Letters. 296 (2), 257-265 (2009).
- Yamaguchi, M., et al. Structome of Saccharomyces cerevisiae determined by freeze-substitution and serial ultrathin sectioning electron microscopy. Journal of Electron Microscopy. 60 (5), 321-335 (2011).
- Yamada, H., et al. Structome analysis of Escherichia coli cells by serial ultrathin sectioning reveals the precise cell profiles and the ribosome density. Microscopy. 66 (4), 283-294 (2017).
- Yamaguchi, M., Ohkusu, M., Sameshima, M., Kawamoto, S. Safe specimen preparation for electron microscopy of pathogenic fungi by freeze-substitution after glutaraldehyde fixation. Japanese Journal of Medical Mycology. 46 (3), 187-192 (2005).
- Yamaguchi, M., et al. Improved preservation of fine structure of deep-sea microorganisms by freeze-substitution after glutaraldehyde fixation. Journal of Electron Microscopy. 60 (4), 283-287 (2011).
- Yamaguchi, M., et al. Prokaryote or eukaryote? A unique microorganism from the deep-sea. Journal of Electron Microscopy. 61 (6), 423-431 (2012).
- Yamada, H., Chikamatsu, K., Aono, A., Mitarai, S. Pre-fixation of virulent Mycobacterium tuberculosis with glutaraldehyde preserves exquisite ultrastructure on transmission electron microscopy through cryofixation and freeze-substitution with osmium-acetone at ultralow temperature. Journal of Microbiological Methods. 96, 50-55 (2014).
- Yamaguchi, M. An electron microscopic study of microorganisms: from influenza virus to deep-sea microorganisms. JSM Mycotoxins. 65 (2), 81-99 (2015).
- Yamaguchi, M., et al. High-voltage electron microscopy tomography and structome analysis of unique spiral bacteria from the deep sea. Microscopy. 65 (4), 363-369 (2016).
- Yamaguchi, M., Yamada, H., Uematsu, K., Horinouchi, Y., Chibana, H. Electron microscopy and structome analysis of unique amorphous bacteria from the deep sea. Cytologia. 83 (3), 337-342 (2018).
- Yamaguchi, M., Yamada, H., Chibana, H. Deep-sea bacteria harboring bacterial endosymbionts in a cytoplasm?: 3D electron microscopy by serial ultrathin sectioning of freeze-substituted specimen. Cytologia. 85 (3), 209-211 (2020).
- Yamaguchi, M., Takahashi-Nakaguchi, A., Aida, Y., Sato-Okamoto, M., Chibana, H. Convenient method for better preservation of fine structures of cultured macrophages and engulfed yeast cells by freeze-substitution fixation. Microscopy. 66 (3), 209-211 (2017).
- Aoki, S., et al. Shift in energy metabolism caused by glucocorticoids enhances the effect of cytotoxic anticancer drugs against acute lymphoblastic leukemia cells. Oncotarget. 8 (55), 94271-94285 (2017).
- Hirao, T., et al. Altered intracellular signaling by imatinib increases the anticancer effects of tyrosine kinase inhibitors in chronic myelogenous leukemia cells. Cancer Science. 109 (1), 121-131 (2018).
- Yamaguchi, M., et al. Sandwich freezing device for rapid freezing of viruses, bacteria, yeast, cultured cells, and animal and human tissues in electron microscopy. Microscopy. 70 (2), 215-223 (2021).
- Yamaguchi, M. Troubleshooting in specimen preparation of microorganisms. Kenbikyo. 42, 26-28 (2007).
- Yamaguchi, M., Aoyama, T., Yamada, N., Chibana, H. Quantitative measurement of hydrophilicity/hydrophobicity of the plasma-polymerized naphthalene film (Super support film) and other support films and grids in electron microscopy. Microscopy. 65 (55), 444-450 (2016).
- Yamaguchi, M., Chibana, H. A method for obtaining serial ultrathin sections of microorganisms in transmission electron microscopy. Journal of Visualized Experiments: JoVE. (131), e56235 (2018).
- Yamaguchi, M., Shimizu, M., Yamaguchi, T., Ohkusu, M., Kawamoto, S. Repeated use of uranyl acetate solution for section staining in transmission electron microscopy. Plant Morphology. 17 (1), 57-59 (2005).
- Yamaguchi, M., Tanaka, A., Suzuki, T. A support film of plasma-polymerized naphthalene for electron microscopy: method of preparation and application. Journal of Electron Microscopy. 41 (1), 7-13 (1992).
- Yamaguchi, M., et al. Cryo-electron microscopy of hepatitis B virus core particles produced by transformed yeast: comparison with negative staining and ultrathin sectioning. Journal of Electron Microscopy. 37 (6), 337-341 (1988).
- Yamaguchi, M., et al. Dynamics of hepatitis B virus core antigen in a transformed yeast cell: analysis with an inducible system. Journal of Electron Microscopy. 43 (6), 386-393 (1994).
- Yamaguchi, M., Miyatsu, T., Mizokami, H., Matsuoka, L., Takeo, K. Translocation of hepatitis B virus core particles through nuclear pores in transformed yeast cells. Journal of Electron Microscopy. 45 (4), 321-324 (1996).
- Sipiczki, M., Takeo, K., Yamaguchi, M., Yoshida, S., Miklos, I. Environmentally controlled dimorphic cycle in fission yeast. Microbiology. 144, 1319-1330 (1998).
- Sipiczki, M., et al. Role of cell shape in determination of the division plane in Schizosaccharomyces pombe: random orientation of septa in spherical cells. Journal of Bacteriology. 182 (6), 1693-1701 (2000).
- Encz, i. K., Yamaguchi, M., Sipiczki, M. Morphology transition genes in the dimorphic fission yeast Schizosaccharomyces japonicus. Antonie van Leeuwenhoek. 92 (2), 143-154 (2007).
- Kopecka, M., et al. Microtubules and actin cytoskeleton in Cryptococcus neoformans compared with ascomycetous budding and fission yeasts. European Journal of Cell Biology. 80 (4), 303-311 (2001).
- Yamaguchi, M., Biswas, S. K., Kita, S., Aikawa, E., Takeo, K. Electron microscopy of pathogenic yeasts Cryptococcus neoformans and Exophiala dermatitidis by high-pressure freezing. Journal of Electron Microscopy. 51 (1), 21-27 (2002).
- Ikeda, R., et al. Contribution of the mannan backbone of cryptococcal glucuroxylomannan and a glycolytic enzyme of Staphylococcus aureus to contact-mediated killing of Cryptococcus neoformans. Journal of Bacteriology. 189 (13), 4815-4826 (2007).
- Yamaguchi, M., et al. The spindle pole body of the pathogenic yeast Cryptococcus neoformans: variation in morphology and positional relationship to the nucleolus and the bud in interphase cells. Journal of Electron Microscopy. 59 (2), 165-172 (2010).
- Kozubowski, L., et al. Ordered kinetochore assembly in the human pathogenic basidiomycetous yeast Cryptococcus neoformans. MBio. 4 (5), 00614 (2013).
- Stepanova, A. A., Yamaguchi, M., Chibana, H., Vasilyeva, N. V. Ultrastructural aspects of cell components migration during budding in the yeast Cryptococcus leurentii. Problems in Medical Mycology. 18 (3), 24-29 (2016).
- Ohkusu, M., et al. Cellular and nuclear characteristics of Exophiala dermatitidis. Studies in Mycology. 43 (43), 143-150 (1999).
- Yamaguchi, M., Biswas, S. K., Suzuki, Y., Furukawa, H., Takeo, K. Three-dimensional reconstruction of a pathogenic yeast Exophiala dermatitidis cell by freeze-substitution and serial sectioning electron microscopy. FEMS Microbiology Letters. 219 (1), 17-21 (2003).
- Yamaguchi, M., et al. The spindle pole body of the pathogenic yeast Exophiala dermatitidis: variation in morphology and positional relationship to the nucleolus and the bud in interphase cells. European Journal of Cell Biology. 82 (10), 531-538 (2003).
- Biswas, S. K., Yamaguchi, M., Naoe, N., Takashima, T., Takeo, K. Quantitative three-dimensional structural analysis of Exophiala dermatitidis yeast cells by freeze-substitution and serial ultrathin sectioning. Journal of Electron Microscopy. 52 (2), 133-143 (2003).
- Takaya, N., et al. Cytochrome P450nor, a novel class of mitochondrial cytochrome P450 involved in nitrate respiration in the fungus Fusarium oxysporum. Archives of Biochemistry and Biophysics. 372 (2), 340-346 (1999).
- Zhou, Z., et al. Ammonia fermentation, a novel anoxic metabolism of nitrate by fungi. Journal of Biological Chemistry. 277 (3), 1892-1896 (2002).
- Takasaki, K., et al. Fungal ammonia fermentation, a novel metabolic mechanism that couples the dissimilatory and assimilatory pathways of both nitrate and ethanol. Role of acetyl CoA synthetase in anaerobic ATP synthesis. Journal of Biological Chemistry. 279 (13), 12414-12420 (2004).
- Kopecka, M., et al. Analysis of microtubules and F-actin structures in hyphae and conidia development in opportunistic human pathogenic black yeast Aureobasidium pullulans. Microbiology. 149, 865-876 (2003).
- Ueno, K., Namiki, Y., Mitani, H., Yamaguchi, M., Chibana, H. Differential cell wall remodeling of two chitin synthase deletants Δchs3A and Δchs3B in the pathogenic yeast Candida glabrata. FEMS Yeast Research. 11 (5), 398-407 (2011).
- Ikezaki, S., et al. Mild heat stress affects on the cell wall structure in Candida albicans biofilm. Medical Mycology Journal. 60 (2), 29-37 (2019).
- Gabriel, M., et al. The cytoskeleton in the unique cell reproduction by conidiogenesis of the long-neck yeast Fellomyces (Sterigmatomyces) fuzhouensis. Protoplasma. 229 (1), 33-44 (2006).
- Yoshimi, A., et al. Functional analysis of the α-1,3-glucan synthase genes agsA and agsB in Aspergillus nidulans: agsB is the major α-1,3-glucan synthase in this fungus. PLoS One. 8 (1), 54893 (2013).
- Yamada, H., Mitarai, S., Chikamatsu, K., Mizuno, K., Yamaguchi, M. Novel freeze-substitution electron microscopy provides new aspects of virulent Mycobacterium tuberculosis with visualization of the outer membrane and satisfying biosafety requirements. Journal of Microbiological Methods. 80 (1), 14-18 (2010).
- Yamada, H., Yamaguchi, M., Chikamatsu, K., Aono, A., Mitarai, S. Structome analysis of virulent Mycobacterium tuberculosis, which survives with only 700 ribosomes at density per 0.1 fl cytoplasm. PLoS One. 10 (1), 0117109 (2015).
- Shiratori, R., et al. Glycolytic suppression dramatically changes the intracellular metabolic profile of multiple cancer cell lines in a mitochondrial metabolism-dependent manner. Scientfic Reports. 9 (1), 18699 (2019).
- McDonald, K. L. Out with the old and in with the new: rapid specimen preparation procedures for electron microscopy of sectioned biological material. Protoplasma. 251 (2), 429-448 (2014).
- Small, J. V. Organization of actin in the leading edge of cultured cells: influence of osmium tetroxide and dehydration on the ultrastructure of actin meshworks. Journal of Cell Biology. 91 (3), 695-705 (1981).
- Ohno, S., Terada, N., Fujii, Y., Ueda, H., Takayama, I. Dynamic structure of glomerular capillary loop as revealed by an in vivo cryotechnique. Virchows Archiv. 427 (5), 519-527 (1996).

