Bioprospecting of Extremophilic Microorganisms to Address Environmental Pollution
Summary
The isolation of heavy metal-resistant microbes from geothermal springs is a hot topic for the development of bioremediation and environmental monitoring biosystems. This study provides a methodological approach for isolating and identifying heavy metal tolerant bacteria from hot springs.
Abstract
Geothermal springs are rich in various metal ions due to the interaction between rock and water that takes place in the deep aquifer. Moreover, due to seasonality variation in pH and temperature, fluctuation in element composition is periodically observed within these extreme environments, influencing the environmental microbial communities. Extremophilic microorganisms that thrive in volcanic thermal vents have developed resistance mechanisms to handle several metal ions present in the environment, thus taking part to complex metal biogeochemical cycles. Moreover, extremophiles and their products have found an extensive foothold in the market, and this holds true especially for their enzymes. In this context, their characterization is functional to the development of biosystems and bioprocesses for environmental monitoring and bioremediation. To date, the isolation and cultivation under laboratory conditions of extremophilic microorganisms still represent a bottleneck for fully exploiting their biotechnological potential. This work describes a streamlined protocol for the isolation of thermophilic microorganisms from hot springs as well as their genotypical and phenotypical identification through the following steps: (1) Sampling of microorganisms from geothermal sites (“Pisciarelli”, a volcanic area of Campi Flegrei in Naples, Italy); (2) Isolation of heavy metal resistant microorganisms; (3) Identification of microbial isolates; (4) Phenotypical characterization of the isolates. The methodologies described in this work might be generally applied also for the isolation of microorganisms from other extreme environments.
Introduction
The extreme environments on our planet are excellent sources of microorganisms capable of tolerating harsh conditions (i.e., temperature, pH, salinity, pressure, and heavy metals)1,2, being Iceland, Italy, USA, New Zealand, Japan, Central Africa and India, the best-recognized and studied volcanic areas3,4,5,6,7,8,9. Thermophiles have evolved in harsh environments in a range of temperatures from 45 °C to 80 °C10,11,12. Thermophilic microorganisms, either belonging to the archaeal or bacterial kingdoms, are a reservoir for the study of biodiversity, phylogenesis, and the production of exclusive biomolecules for industrial applications 13,14,15,16. Indeed, in the last decades, the continuous industrial demand in the global market has encouraged the exploitation of extremophiles and thermozymes for their diversified applications in several biotechnological fields 17,18,19.
Hot springs, where organisms live in consortia, are rich sources of biodiversity, thus representing an attractive habitat to study microbial ecology20,21. Moreover, these volcanic metal-rich areas are commonly colonized by microorganisms that have evolved tolerance systems to survive and adapt to the presence of heavy metals22,23 and are therefore actively involved in their biogeochemical cycles. Nowadays, heavy metals are considered priority pollutants for humans and the environment. The heavy-metal-resistant microorganisms are able to solubilize and precipitate metals by transforming them and remodeling their ecosystems24,25. The comprehension of the molecular mechanisms of heavy-metal resistance is a hot topic for the urgency to develop novel green approaches26,27,28. In this context, the discovery of new tolerant bacteria represents the starting point for developing new strategies for environmental bioremediation24,29. In accompanying the efforts to explore hydrothermal environments through microbiological procedures and increase knowledge on the role of the gene(s) underpinning heavy metal tolerance, a microbial screening was conducted in the hot-spring area of Campi Flegrei in Italy. This heavy metal-rich environment shows a powerful hydrothermal activity, fumarole, and boiling pools, variable in pH and temperature in dependence of seasonality, rainfall, and underground geological movements30. In this perspective, we describe an easy-to-apply and efficacious way to isolate bacteria resistant to heavy metals, for example, Geobacillus stearothermophilus GF1631 (named as isolate 1) and Alicyclobacillus mali FL1832 (named as isolate 2) from Pisciarelli area of Campi Flegrei.
Protocol
1. Sampling of microorganisms from geothermal sites
- Choose the site for sampling using as criterion places with desired temperature and pH. Measure the physical parameters through a digital thermocouple probe, inserting it into the selected pools or muds.
- Collect 20 g of soils samples (in this case, from mud in the hydrothermal site of Pisciarelli Solfatara), picking them up with a sterilized spoon. Take at least two samples for each site chosen.
- Put the samples in 50 mL sterile polypropylene tubes and immediately close.
- Measure pH and temperature with a digital thermocouple probe by directly inserting it into the sampling site. After use, rinse the probe carefully with deionized water.
2. Isolation of heavy metal resistant microorganisms
NOTE: Perform steps 2.1-2.7 under a sterile biological hood.
- Inoculate 2 g of each collected sample into 50 mL of freshly prepared Luria-Bertani medium (LB), in which the pH has been adjusted to 4 or 7 through the addition of HCl or NaOH.
- Incubate the samples at the same temperature of the sampling site and at ±5 °C (55 °C and 60 °C for Pisciarelli samples) in a temperature-controlled orbital shaker for 24 h with a shaking rate of 180 rpm.
- Plate 200 µL of the grown samples on LB agar (pH 4 or pH 7) and incubate in static condition for 48 h at 55 °C or 60 °C.
- Isolate single colonies and repeat streak-plating cycles (steps 2.3 and 2.4) at least three times.
- To prepare -80 °C frozen cell stocks, grow the cultures overnight (ON) and add to the grown cells 20% glycerol (in a final volume of 1 mL); use a mixture of acetone and dry ice for fast freezing.
- To prepare an inoculum from a glycerol stock, inoculate 50 μL in 50 mL of LB (pH 4 or pH 6) and incubate at 55 °C or 60 °C in the orbital shaker at 180 rpm ON.
- To obtain a growth profile, dilute a preculture (obtained from step 2.6) to 0.1 OD600 nm in 10 mL of LB (pH 4 or pH 6), grow the cells at 55 °C or 60 °C for 16 h in the orbital shaker, and measure the OD600 nm at 30 min intervals.
- Construct a growth curve from the data obtained in step 2.7 with time (min) on X-axis and OD600 nm on Y-axis.
- Realize the same growth curve described in steps 2.7 and 2.8 but varying the pH (± 1 unit) of the culture medium (e.g., pH 3 and 5 for samples grown at pH 4) to determine the optimal pH for laboratory conditions.
3. Identification of microbial isolates
- Preparation of genomic DNA
- Inoculate the isolate streaked from the glycerol stock in 50 mL of LB medium (pH 4 or pH 6) and grow in an orbital shaker at 55 °C or 60 °C at 180 rpm ON.
- Harvest the ON culture by centrifugation for 10 min at 5000 x g. Discard the supernatant.
- Prepare 10 mL of bacteria lysis buffer composed by: 20 mM Tris-HCl pH 8.0, 2 mM EDTA, 1.2% Triton X-100, and lysozyme (20 mg/mL) immediately before use.
- Resuspend the pellet in 180 µL of bacteria lysis buffer. Incubate for 30 min at 37 °C.
- Follow the guidelines indicated by a Genomic DNA Purification kit (Table of Materials) to extract genomic DNA.
- Quantify the extracted genomic DNA and its purity by UV-Vis measurement. For purity determine ratios-OD 260/280 nm and OD 260/230 nm.
- Assess the integrity of the genomic DNA by loading 200 ng of each sample on a 0.8% agarose gel and comparing the size distribution to a high-weight molecular marker.
- Commission to an external service the 16S rRNA fragment preparation, sequencing, and comparative analysis of the sequence obtained (1000 bp) with those present in the nucleotide database of the US National Center for Biotechnology Information (NCBI)33.
- To corroborate data of 16S rRNA sequencing, also perform automated ribotyping on the digested chromosomal DNA (external service, Table of Materials).
- In the case in which the specie identification cannot be determined only with ribotyping data, commission a MALDI-TOF MS analysis for fatty acid identification.
- To perform a phylogenetic analysis of the genus identified, analyze the 16S rRNA sequence of the isolate with BLASTn34. Sequences with identities from 99% to 97% must be used to build a multiple sequence alignment using CLUSTAL Omega35. Construct a neighbor-joining tree using the default option of ClustalW2 (Simple Phylogeny).
4. Heavy-metals and antibiotics susceptibility
- Inoculate the isolate from a glycerol stock (see step 2.5) and grow it in 200 mL of LB under the optimal pH and temperature conditions previously determined.
- Dilute each preculture at 0.1 OD600 nm in 5 mL of LB medium (at the appropriate pH) containing increasing concentrations of heavy metals. The concentrations vary from 0.01-120 mM for heavy metals [As(V), As(III), Cd(II), Co(III), Cr(VI), Cu(II), Hg(II), Ni(II), V(V)] or 0.5-1 mg/mL for the antibiotics [Ampicillin, Bacitracin, Chloramphenicol, Ciprofloxacin, Erythromycin, Kanamycin, Streptomycin, Tetracycline, and Vancomycin].
- Perform heavy metal and antibiotic treatments separately. Use a 50 mL polypropylene tube and grow the cells in a temperature-controlled orbital shaker with a shaking rate of 180 rpm at 55 °C or 60°C for 16 h for each condition/treatment.
- Calculate Minimum Inhibitory Concentration (MIC) either for antibiotics or heavy metals by identifying the concentration values in the tubes where microbial growth does not occur, i.e., determining the values that completely inhibit cell growth after 16 h.
- Check that the concentration is inhibitory and not lethal for the cells by plating 200 μL of the culture grown at the value that is considered as MIC on LB-agar plates (at the appropriate pH and temperature) and verifying the presence of colonies after ON incubation.
NOTE: Since the culture on LB agar plate is viable at 4 °C only for a few weeks, in order to preserve the isolates for a longer time, glycerol stocks were prepared and stored at -80 °C. For MIC determination, at least three independent replicates using independent cultures were carried out. The standard deviation was calculated among triplicate experiments.
Representative Results
Sampling site
This protocol illustrates a method for the isolation of heavy metal-resistant bacteria from a hot spring. In this study, the Pisciarelli area, an acid-sulfidic geothermal environment, was used as a sampling site (Figure 1). This ecosystem is characterized by the flow of aggressive sulfurous fluids derived from volcanic activities. It has been demonstrated that the microbial communities in acid-sulfidic geothermal systems are subjected to extreme selective pressure made by the presence of high concentrations of heavy metals. The samples were collected in two different periods of the year (April and September) from2,21 a mud pool marginal with respect to a bubbling mud pool. In the mud pool, fluctuations in the pH values (~pH 6 in April and ~pH 5 in September) were registered, while the temperature was ~55 °C in both cases. However, higher temperatures were also recorded in the mud pool (~70 °C) in other years32.
Isolation and identification
The collected samples were inoculated in LB medium and incubated for 24 h at 55 °C and 60 °C as reported previously, hence setting the lab conditions for the growth of the cell samples to mimic the environmental chemical-physical conditions. To favor cell growth, single colonies were streaked on the plate and isolated after several dilutions (at least 3) in a rich-liquid medium; the isolated strains showed their optimal growth temperature at 55 °C and 60 °C (Figure 2). To identify the new isolates, a genomic DNA preparation was carried out and 16S rRNA sequencing and fatty acids mass spectrometry analysis was accomplished as an external service. As reported, the analysis of the fatty acids is a powerful bioanalytical method that helps in the precise identification of bacteria when combined with other approaches36. Multiple alignments of 16S rRNA were used to build the phylogenetic tree to identify the closest relatives37.
Heavy metal susceptibility test
The coexistence of toxic molecules characterizes solfataric environments. In particular, hot springs in Pisciarelli are characterized by high levels of CO2, H2S, NH4 in coexistence with As, Hg, Fe, Be, Ni, Co, Cu30,38. For this reason, a phenotypical characterization of the isolated microorganisms was performed in the presence of an increasing concentration of heavy metals, as reported in Table 1. Interestingly, isolate 1 showed higher tolerance to As(V) and V(V). The high resistance to both arsenate and vanadate can be due to their chemical structures; in fact, both ions are similar to the phosphate ions, suggesting that V(V) and As(V) could be taken up by cells through phosphate transport systems. These isolates turned out to be also resistant to Cd(II), although the MIC value was relatively low. This result can be explained by the absence of Cd(II) in the pool. Although the two microorganisms were sampled in the same site, they showed different heavy metal resistance profiles. However, they were sampled in different periods, thus pointing to the season-dependent variation in the heavy metals concentration as the main driving force shaping the composition of the microbial communities and their differential resistance to heavy metals39. From this comparative data, it has been shown that isolate 1 has a strong resistance to As(V), while isolate 2 for As(III). Further genetic investigations are required to unravel the molecular resistance mechanisms and better understand how the phenotypes are affected by the selective pressure of hot springs.
Antibiotics resistance tests
The microbial strains evolved in extreme environments usually exhibit resistance to different antibiotics. The correlation between the heavy metal resistance and antibiotics is well-known40. For this reason, we tested the resistance to antibiotics for both isolates (Table 2). Isolate 1 showed high sensitivity to all the tested antibiotics, even when low concentrations were used. In contrast, isolate 2 is resistant to all the antibiotics tested, with the exception of chloramphenicol and tetracycline. Interestingly, the determined MIC values towards ampicillin, erythromycin, kanamycin, streptomycin, and vancomycin were comparable to those of other antibiotic-resistant bacteria and even higher for bacitracin and ciprofloxacin41. These fascinating data deserve further investigations; probably, due to random mutations or horizontal gene transfer, the microorganism has acquired antibiotic resistance, which could represent a selective advantage in such extreme environmental conditions.

Figure 1. Sampling site: solfataric area of Pisciarelli, Campi Flegrei (Naples, Italy). The sampling site is located at 40° 49' 45.3" N – 14° 08' 49.9 E, in the geothermal area of Pisciarelli fumarole. Please click here to view a larger version of this figure.
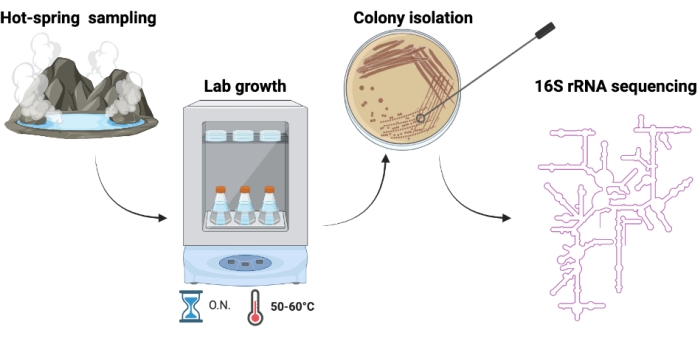
Figure 2. Schematic representation of the experimental procedure. Microorganisms are sampled in hot springs, cultivated in the laboratory, isolated through repeated streaking and plating, and genotypically identified upon 16S rRNA sequencing. Please click here to view a larger version of this figure.
| Metal ions | Isolate 1 | Isolate 2 |
| As (III) | 1.9 mM | 41 mM |
| As (V) | 117 mM | 11 mM |
| Cd (II) | 0.9 mM | 0.8 µM |
| Co (II) | 2 mM | 3 mM |
| Co (III) | 2.75 mM | n.a. |
| Cr (VI) | 0.25 mM | n.a. |
| Cu (II) | 4.1 mM | 0.5 mM |
| Hg (II) | 20 µM | 17 µM |
| Ni (II) | 1.3 mM | 30 mM |
| V (V) | 128 mM | n.a |
Table 1. MIC values towards heavy metal ions of the isolates. MICs are considered as the minimal concentration values that completely inhibit cell growth after 16 h; the values are reported as average of three experiments.
| Antibiotics | Isolate 1 | Isolate 2 |
| Ampicillin | n.d. | 20 µg/mL |
| Bacitracin | n.d. | 700 µg/mL |
| Chloramphenicol | n.d. | <0.5 µg/mL |
| Ciprofloxacin | n.d. | >1 mg/mL |
| Erythromycin | n.d. | 70 µg/mL |
| Kanamycin | n.d. | 80 µg/mL |
| Streptomycin | n.d. | 70 µg/mL |
| Tetracycline | n.d. | <0.5 µg/mL |
| Vancomycin | n.d. | 1 µg/mL |
Table 2. MIC values towards antibiotics of the isolates. MICs are considered as the minimal concentrations that completely inhibit the cell growth after 16 h; the values are reported as average of three experiments.
Discussion
Hot springs contain an untapped diversity of microbiomes with equally diverse metabolic capacities12. The development of strategies for the isolation of microorganisms that can efficiently convert heavy metals into less toxic compounds10 represents a research area of growing interest worldwide. This paper aims to describe a streamlined approach for the screening and isolation of microbes with the ability to resist toxic chemicals. The method described can be easily modified to isolate microbes from diverse environmental sources such as water, food, soil, or sediment. However, there are some limitations in this technique related to the reliance on microbial culturing. Therefore, this setup would not be suitable for isolating bacteria from an environment that is not easily culturable. One way to overcome this issue is to use different bacterial media (i.e., selective media or pre-adaptation strategies) and longer incubation times42.
Nevertheless, the majority of species of interest for bioremediation are expected to grow under the conditions described herein. This protocol has some advantages over traditional plating techniques, considering that selective agar media for chemicals are unknown so far. The use of MIC to identify resistant microbes is a quick strategy to be exploited on individual isolates that opens the way to the characterization of new species or new strains. This study demonstrates the usefulness of such a method to select environmental microorganisms that can contribute to effective bioremediation by inactivating the pollutants and converting them into harmless products.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was supported by ERA-NET Cofund MarTERA: "FLAshMoB: Functional Amyloid Chimera for Marine Biosensing", PRIN 2017-PANACEA CUP:E69E19000530001 and by GoodbyWaste: ObtainGOOD products-exploit BY-products-reduce WASTE, MIUR 2017-JTNK78.006, Italy. We thank Dr. Monica Piochi and Dr. Angela Mormone (Istituto Nazionale di Geofisica e Vulcanologia, Sezione di Napoli Osservatorio Vesuviano, Italy) for the identification and characterization of geothermal site.
Materials
| Ampicillin | Sigma Aldrich | A9393 | |
| Aura Mini | bio air s.c.r.l. | Biological hood | |
| Bacitracin | Sigma Aldrich | B0125 | |
| Cadmium chloride | Sigma Aldrich | 202908 | |
| Chloramphenicol | Sigma Aldrich | C0378 | |
| Ciprofloxacin | Sigma Aldrich | 17850 | |
| Cobalt chloride | Sigma Aldrich | C8661 | |
| Copper chloride | Sigma Aldrich | 224332 | |
| Erythromycin | Sigma Aldrich | E5389 | |
| Exernal Service | DSMZ | Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures GmbH | |
| Genomic DNA Purification Kit | Thermo Scientific | #K0721 | |
| Kanamycin sulphate | Sigma Aldrich | 60615 | |
| MaxQTM 4000 Benchtop Orbital Shaker | Thermo Scientific | SHKE4000 | |
| Mercury chloride | Sigma Aldrich | 215465 | |
| NanoDrop 1000 Spectrophotometer | Thermo Scientific | ||
| Nickel chloride | Sigma Aldrich | 654507 | |
| Orion Star A221 Portable pH Meter | Thermo Scientific | STARA2218 | |
| Sodium (meta) arsenite | Sigma Aldrich | S7400 | |
| Sodium arsenate dibasic heptahydrate | Sigma Aldrich | A6756 | |
| Sodium chloride | Sigma Aldrich | S5886 | |
| Streptomycin | Sigma Aldrich | S6501 | |
| Tetracycline | Sigma Aldrich | 87128 | |
| Tryptone BioChemica | Applichem Panreac | A1553 | |
| Vancomycin | Sigma Aldrich | PHR1732 | |
| Yeast extract for molecular biology | Applichem Panreac | A3732 |
Referências
- Arora, N. K., Panosyan, H. Extremophiles: applications and roles in environmental sustainability. Environmental Sustainability. 2, 217-218 (2019).
- Gallo, G., Puopolo, R., Carbonaro, M., Maresca, E., Fiorentino, G. Extremophiles, a nifty tool to face environmental pollution: From exploitation of metabolism to genome engineering. International Journal of Environmental Research and Public Health. 18 (10), 5228 (2021).
- Saxena, R., et al. Metagenomic analysis of hot springs in Central India reveals hydrocarbon degrading thermophiles and pathways essential for survival in extreme environments. Frontiers in Microbiology. 7, 2123 (2017).
- Papke, R. T., Ramsing, N. B., Bateson, M. M., Ward, D. M. Geographical isolation in hot spring cyanobacteria. Environmental Microbiology. 5 (8), 650-659 (2003).
- Zitelle, L., Lan Pe, N. I. al The role of photosynthesis and CO2 evasion in travertine formation: a quantitative investigation at an important travertine-depositing hot spring. Journal of the Geological Society. 164, 843-853 (2007).
- Kubo, K., Knittel, K., Amann, R., Fukui, M., Matsuura, K. Sulfur-metabolizing bacterial populations in microbial mats of the Nakabusa hot spring. Japan. Systematic and Applied Microbiology. 34 (4), 293-302 (2011).
- Siljeström, S., Li, X., Brinckerhoff, W., van Amerom, F., Cady, S. L. ExoMars mars organic molecule analyzer (MOMA) laser desorption/ionization mass spectrometry (LDI-MS) analysis of phototrophic communities from a silica-depositing hot spring in Yellowstone national park, USA. Astrobiology. , (2021).
- Aulitto, M., Tom, L. M., Ceja-Navarro, J. A., Simmons, B. A., Singer, S. W. Whole-genome sequence of Brevibacillus borstelensis SDM, isolated from a sorghum-adapted microbial community. Microbiology Resource Announcements. 9 (48), 8-9 (2020).
- Antranikian, G., et al. Diversity of bacteria and archaea from two shallow marine hydrothermal vents from Vulcano Island. Extremophiles. 21 (4), 733-742 (2017).
- Gallo, G., Puopolo, R., Limauro, D., Bartolucci, S., Fiorentino, G. Metal-tolerant thermophiles: from the analysis of resistance mechanisms to their biotechnological exploitation. The Open Biochemistry Journal. 12 (1), 149-160 (2018).
- Aulitto, M., et al. Draft genome sequence of Bacillus coagulans MA-13, a thermophilic lactic acid producer from lignocellulose. Microbiology Resource Announcements. 8 (23), 341-360 (2019).
- Mehta, D., Satyanarayana, T. Diversity of hot environments and thermophilic microbes. Thermophilic Microbes in Environmental and Industrial Biotechnology: Biotechnology of Thermophiles. , (2013).
- Fusco, S., et al. The interaction between the F55 virus-encoded transcription regulator and the RadA host recombinase reveals a common strategy in Archaea and Bacteria to sense the UV-induced damage to the host DNA. Biochimica et Biophysica Acta – Gene Regulatory Mechanisms. 1863 (5), (2020).
- Puopolo, R., et al. Self-assembling thermostable chimeras as new platform for arsenic biosensing. Scientific Reports. 11 (1), (2021).
- Fiorentino, G., Contursi, P., Gallo, G., Bartolucci, S., Limauro, D. A peroxiredoxin of Thermus thermophilus HB27: Biochemical characterization of a new player in the antioxidant defence. International Journal of Biological Macromolecules. 153, 608-615 (2020).
- Fiorentino, G., Del Giudice, I., Bartolucci, S., Durante, L., Martino, L., Del Vecchio, P. Identification and physicochemical characterization of BldR2 from Sulfolobus solfataricus, a novel archaeal member of the MarR transcription factor family. Bioquímica. 50 (31), 6607-6621 (2011).
- Bhattacharya, A., Gupta, A. G. . Microbial Extremozymes. Current trends in applicability of thermophiles and thermozymes in bioremediation of environmental pollutants. , 161-176 (2022).
- Aulitto, M., et al. Prebiotic properties of Bacillus coagulans MA-13: Production of galactoside hydrolyzing enzymes and characterization of the transglycosylation properties of a GH42 β-galactosidase. Microbial Cell Factories. 20 (1), 1-18 (2021).
- Ing, N., et al. A multiplexed nanostructure-initiator mass spectrometry (NIMS) assay for simultaneously detecting glycosyl hydrolase and lignin modifying enzyme activities. Scientific Reports. 11 (1), 11803 (2021).
- Saw, J. H. W. Characterizing the uncultivated microbial minority: towards understanding the roles of the rare biosphere in microbial communities. mSystems. 6 (4), 0077321 (2021).
- He, Q., et al. Temperature and microbial interactions drive the deterministic assembly processes in sediments of hot springs. Science of the Total Environment. 772, 145465 (2021).
- Shakhatreh, M. A. K., et al. Microbiological analysis, antimicrobial activity, and heavy-metals content of Jordanian Ma’in hot-springs water. Journal of Infection and Public Health. 10 (6), 789-793 (2017).
- Antonucci, I., et al. An ArsR/SmtB family member regulates arsenic resistance genes unusually arranged in Thermus thermophilus HB27. Microbial Biotechnology. 10 (6), 1690-1701 (2017).
- Ozdemir, S., Kılınç, E., Poli, A., Nicolaus, B. Biosorption of Heavy Metals (Cd 2+, Cu 2+ , Co 2+ , and Mn 2+ ) by Thermophilic Bacteria, Geobacillus thermantarcticus and Anoxybacillus amylolyticus Equilibrium and Kinetic Studies. Bioremediation Journal. 17 (2), 86-96 (2013).
- Hlihor, R. -. M., Apostol, L. -. C., Gavrilescu, M. Environmental bioremediation by biosorption and bioaccumulation: Principles and applications. Enhancing Cleanup of Environmental Pollutants: Volume 1: Biological Approaches. , 289-315 (2017).
- Del Giudice, I., Limauro, D., Pedone, E., Bartolucci, S., Fiorentino, G. A novel arsenate reductase from the bacterium Thermus thermophilus HB27: its role in arsenic detoxification. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 1834 (10), 2071-2079 (2013).
- Politi, J., Spadavecchia, J., Fiorentino, G., Antonucci, I., Casale, S., De Stefano, L. Interaction of Thermus thermophilus ArsC enzyme and gold nanoparticles naked-eye assays speciation between As(III) and As(V). Nanotechnology. 26 (43), 435703 (2015).
- Antonucci, I., et al. Characterization of a promiscuous cadmium and arsenic resistance mechanism in Thermus thermophilus HB27 and potential application of a novel bioreporter system. Microbial Cell Factories. 17 (1), (2018).
- Ilyas, S., Lee, J. C., Kim, B. S. Bioremoval of heavy metals from recycling industry electronic waste by a consortium of moderate thermophiles: Process development and optimization. Journal of Cleaner Production. 70, 194-202 (2014).
- Piochi, M., Mormone, A., Strauss, H., Balassone, G. The acid-sulfate zone and the mineral alteration styles of the Roman Puteolis (Neapolitan area, Italy): clues on fluid fracturing progression at the Campi Flegrei volcano. Solid Earth. 10 (6), 1809-1831 (2019).
- Puopolo, R., et al. Identification of a new heavy-metal-resistant strain of Geobacillus stearothermophilus isolated from a hydrothermally active volcanic area in southern Italy. International Journal of Environmental Research and Public Health. 17 (8), 2678 (2020).
- Aulitto, M., et al. Genomic insight of Alicyclobacillus mali FL18 isolated from an Arsenic-rich hot spring. Frontiers in Microbiology. 12, 639697 (2021).
- Agarwala, R., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 46, 8-13 (2018).
- Altschul, S. F., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 25 (17), 3389-3402 (1997).
- Sievers, F., Higgins, D. G. Clustal Omega. Current Protocols in Bioinformatics. 2014, 1-16 (2014).
- Kliem, M., Sauer, S. The essence on mass spectrometry based microbial diagnostics. Current Opinion in Microbiology. 15 (3), 397-402 (2012).
- Madeira, F., et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Research. 47, 636-641 (2019).
- Piochi, M., Mormone, A., Balassone, G., Strauss, H., Troise, C., De Natale, G. Native sulfur, sulfates and sulfides from the active Campi Flegrei volcano (southern Italy): Genetic environments and degassing dynamics revealed by. Journal of Volcanology and Geothermal Research. 301, 180-193 (2015).
- Hsu, H. -. C., et al. Assessment of temporal effects of a mud volcanic eruption on the bacterial community and their predicted metabolic functions in the mud volcanic sites of Niaosong, Southern Taiwan. Nicroorganisms. 9 (11), 2315 (2021).
- Ye, J., Rensing, C., Su, J., Zhu, Y. G. From chemical mixtures to antibiotic resistance. Journal of Environmental Sciences (China). 62, 138-144 (2017).
- Farias, P., et al. Natural hot spots for gain of multiple resistances: arsenic and antibiotic resistances in heterotrophic, aerobic bacteria from marine hydrothermal vent fields. Applied and Environmental Microbiology. 81 (7), 2534-2543 (2015).
- Aulitto, M., Fusco, S., Nickel, D. B., Bartolucci, S., Contursi, P., Franzén, C. J. Seed culture pre-adaptation of Bacillus coagulans MA-13 improves lactic acid production in simultaneous saccharification and fermentation. Biotechnology for Biofuels. 12 (1), 45 (2019).

