Teasing Out the Interplay Between Natural Killer Cells and Nociceptor Neurons
Summary
Nociceptor neurons and NK cells actively interact in an inflammatory context. A co-culture approach enables studying this interplay.
Abstract
Somatosensory neurons have evolved to detect noxious stimuli and activate defensive reflexes. By sharing means of communication, nociceptor neurons also tune host defenses by controlling the activity of the immune system. The communication between these systems is mostly adaptive, helping to protect homeostasis, it can also lead to, or promote, the onset of chronic diseases. Both systems co-evolved to allow for such local interaction, as found in primary and secondary lymphoid tissues and mucosa. Recent studies have demonstrated that nociceptors directly detect and respond to foreign antigens, immune cell-derived cytokines, and microbes.
Nociceptor activation not only results in pain hypersensitivity and itching, but lowers the nociceptor firing threshold, leading to the local release of neuropeptides. The peptides that are produced by, and released from, the peripheral terminals of nociceptors can block the chemotaxis and polarization of lymphocytes, controlling the localization, duration, and type of inflammation. Recent evidence shows that sensory neurons interact with innate immune cells via cell-cell contact, for example, engaging group 2D (NKG2D) receptors on natural killer (NK) cells.
Given that NK cells express the cognate receptors for various nociceptor-produced mediators, it is conceivable that nociceptors use neuropeptides to control the activity of NK cells. Here, we devise a co-culture method to study nociceptor neuron-NK cell interactions in a dish. Using this approach, we found that lumbar nociceptor neurons decrease NK cell cytokine expression. Overall, such a reductionist method could be useful to study how tumor-innervating neurons control the anticancer function of NK cells and how NK cells control the elimination of injured neurons.
Introduction
The cell bodies of sensory neurons originate in the dorsal root ganglia (DRG). The DRG are located in the peripheral nervous system (PNS), between the dorsal horn of the spinal cord and the peripheral nerve terminals. The pseudo-unipolar nature of DRG neurons allows the transfer of information from the peripheral branch, which innervates the target tissue, to the central branch, which carries the somatosensory information to the spinal cord1. Using specialized ion channel receptors, first-order neurons sense threats posed by pathogens, allergens, and pollutants2, leading to the influx of cations (Na+, Ca2+) and the generation of an action potential3,4,5.
These neurons also send antidromic action potential toward the periphery, where the initial danger sensing had occurred, which leads to the local release of neuropeptides1,4. Therefore, the nociceptor neurons serve as a protective mechanism, alerting the host to environmental danger4,5,6,7.
To communicate with second-order neurons, the nociceptors release various neurotransmitters (e.g., glutamate) and neuropeptides (e.g., calcitonin gene-related peptide (CGRP), substance P (SP), and vasoactive intestinal peptide (VIP))6,7.These peptides act on capillaries and promote plasma extravasation, edema, and the local influx and modulation of immune cells2,4,7.
The somatosensory and immune systems utilize a shared communication system composed of cytokines and neuropeptides, and their respective cognate receptors4. While this bidirectional communication helps protect from danger and preserve homeostasis, it can also contribute to disease pathophysiology4.
NK cells are classified as innate lymphoid cells and are specialized to eliminate virally infected cells. NK cell function is governed by a balance of stimulatory and inhibitory receptors, including the activating receptor NKG2D8. The endogenous ligand of NKG2D, retinoic acid early inducible1 (RAE1), is expressed by cells undergoing stress such as tumorigenesis and infection8,9.
Recent investigations have shown that peripheral nerve injury drives sensory neurons to express maladaptive molecules such as stathmin 2 (STMN2) and RAE1. Thus, via cell-cell contact, NKG2D-expressing NK cells were activated by interaction with RAE1-expressing neurons. In turn, NK cells were able to eliminate injured nociceptor neurons and blunt pain hypersensitivity normally associated with nerve injury10. In addition to the NKG2D-RAE1 axis, NK cells express the cognate receptors for various nociceptor-produced mediators. It is therefore possible that these mediators modulate NK cell activity. This paper presents a co-culture method to investigate the biology of the nociceptor neuron-NK cell interaction. This approach will help advance the understanding of how nociceptor neurons modulate innate immune cell responses to injury, infection, or malignancy.
Protocol
The Institutional Animal Care and Use Committees of Université de Montréal (#22053, #22054) approved all animal procedures. See Table 1 for a list of solutions and their composition and the Table of Materials for a list of materials, equipment, and reagents used in this protocol.
1. NK cell isolation, culture, and stimulation
- Generate nociceptor neuron intact (littermate control; TRPV1wt::DTAfl/wt) and ablated (TRPV1cre::DTAfl/wt) mice by crossing lox-stop-lox-diphtheria toxin mice (DTAfl/fl) with TRPV1cre/wt mice.
- Using CO2 inhalation, euthanize a naïve littermate control (TRPV1wt::DTAfl/wt) mouse.
- Collect the spleen in a 1.5 mL microcentrifuge tube containing 500 µL of sterile phosphate-buffered saline (PBS).
- Homogenize the spleen using a pestle.
- Dilute the cells with 1 mL of sterile PBS.
- Filter the mixture through a 50 µm cell strainer into a 15 mL conical tube.
- Top up the volume (10 mL) with sterile PBS.
- Collect 10 µL and count the cells using a hemocytometer.
- Centrifuge the cells (500 × g, 5 min) and remove the supernatant.
- Resuspend the cells at a concentration of 108 cells/mL in supplemented RPMI 1640 medium.
- Follow the manufacturer's instructions to magnetically purify spleen NK cells using a mouse NK cell isolation kit. Confirm NK cell purity using flow cytometry11.
- Collect 10 µL and count the cells using a hemocytometer.
- Centrifuge the cells (500 × g, 5 min) and remove the supernatant.
- Resuspend the NK cells in supplemented RPMI 1640 culture medium (containing IL-2 and IL-15).
- Culture 2 × 106 NK cells/mL in a 96-well U-bottom plate for 48 h.
2. DRG neuron isolation and culture
- At 24 h prior to the end of the NK cell stimulation (step 1.15), coat a 96-well plate with 100 µL/well of laminin (1 ng/mL) and incubate for 45 min (37 °C).
- After incubation, remove the solution and allow the wells to air-dry in a biosafety cabinet.
- Using CO2 inhalation, euthanize nociceptor neuron intact (littermate control; TRPV1wt::DTAfl/wt) or ablated (TRPV1cre::DTAfl/wt) mice.
NOTE: CO2 inhalation is preferred to cervical dislocation as it avoids disrupting the spinal nerves connected to the DRG. - Secure the mouse on the board in a prone position, lift the skin, and incise the skin along the dorsal column. Refer to Perner et al. for a detailed video12.
- Cut off the lumbosacral joint and separate the sacrum from the lumbar spine with scissors.
- Separate the spine by cutting the muscles and ribs on both sides of the spine in the cranial direction until the base of the skull is reached.
- Using scissors, cut off the spine at the atlanto-occipital joint.
- Remove muscle and fatty tissue from the spine.
- Pin and open the vertebral column to remove the spinal cord and gain access to the DRG. Look for the DRG in the intervertebral foramina connected to the spinal cord through the dorsal root but separated from the meninges.
- Harvest the DRGs into a 15 mL conical tube filled with 10 mL of ice-cold supplemented DMEM.
- Centrifuge the preparation (200 × g, 5 min, room temperature) and remove the supernatant.
- Add 250 µL of PBS containing Collagenase IV (1 mg/mL) and Dispase II (2.4 U/mL).
- Incubate the DRG with Collagenase IV/Dispase II (C/D) solution (80 min, 37 °C, mild agitation). When pooling ganglia from several mice, maintain a ratio of 125 µL of C/D solution per ganglion.
- To inactivate the enzymes, add 5 mL of supplemented DMEM medium.
- Centrifuge the solution (200 × g, 5 min) and gently remove the supernatant using a pipette.
- To avoid unwanted cell loss, leave ~100 µL of the supernatant.
- Add 1 mL of supplemented DMEM medium.
- Using a pipette gun and three glass Pasteur pipettes of decreasing sizes, gently triturate (~10 up/down per Pasteur pipette size) the DRG into a single-cell preparation.
- In a biosafety cabinet, dilute bovine serum albumin (BSA) in sterile PBS to a final concentration of 15%. Store 1 mL aliquots at −20 °C.
- Create a BSA gradient in a 15 mL conical tube by adding 2 mL of sterile PBS and slowly dispensing 1 mL of 15% BSA solution at the bottom of the tube. Avoid disrupting the gradient by gently removing the pipette.
- Using a P200 pipette, slowly pipette the triturated ganglia suspension (which contains the neurons) onto the side of the tube into the gradient.
- Centrifuge the neuron-containing BSA gradient (200 × g, 12 min, room temperature). Set the acceleration and deceleration of the centrifuge to the minimum speed.
NOTE: After centrifugation, the neurons will be at the bottom of the tube while the cell debris will be trapped in the BSA gradient. - Gently remove all of the supernatant using a pipette.
- Resuspend the cells in 500 µL of Neurobasal.
- Plate 104 neurons per well (200 µL of neurobasal/well) in a laminin-coated, flat-bottom 96-well plate.
NOTE: On average, an investigator will be able to fill ~8 wells per mouse. - Culture the neurons (37 °C, overnight) to allow attachment.
3. Co-culture and flow cytometry
- Slowly remove the neurobasal from the neuron culture.
- After 48 h stimulation with IL-2 and IL-15 (see steps 1.14-1.15), resuspend the NK cells and add 105 NK cells/well to the neuron culture (96-well flat bottom plate).
NOTE: On average, the investigator will be able to fill ~6 wells per mouse. - Co-culture the cells in supplemented neurobasal MX (37 °C, 48 h).
- Collect the cells, wash with PBS (3x), and centrifuge (500 × g, 5 min, 4 °C).
- Discard the supernatant and stain the cells with Viability Dye eFlour-780 (1:1,000, 15 min, 4 °C).
- Collect the cells, wash with PBS (3x), and centrifuge (500 × g, 5 min, 4 °C).
- Block nonspecific sites on the cells with CD16/32 (1:100, 15 min, 4 °C).
- Collect the cells, wash with PBS (3x), and centrifuge (500 × g, 5 min, 4 °C).
- Stain (15 min, 4 °C) the cells with BV421 anti-NK-1.1 (1:100), fluorescein isothiocyanate (FITC) anti-NKp46 (1:100), and phycoerythrin (PE) anti-GM-CSF (1:100).
- Collect the cells, wash with PBS (3x), and centrifuge (500 × g, 5 min, 4 °C).
- Discard the supernatant, resuspend the cells in FACS buffer (500 µL) and immunophenotype the NK cells by flow cytometry11. See Figure 1A for the gating strategy.
Representative Results
NK cells were magnetically purified from littermate control (TRPV1wt::DTAfl/wt) mice splenocytes and stimulated (48 h) with IL-2 and IL-15. The NK cells were then cultured alone or co-cultured with DRG neurons harvested from nociceptor neuron intact (littermate control; TRPV1wt::DTAfl/wt) or ablated (TRPV1cre::DTAfl/wt) mice. The cells were then exposed to the TRPV1 agonist capsaicin (1 µM) or its vehicle. After 24 h of co-culture, the NK cells were FACS-purified and their activation levels were immunophenotyped using flow cytometry (Figure 1A).
When cultured alone, NK cells expressed basal levels of GM-CSF and NKp46. When co-cultured with DRG neurons harvested from nociceptor intact (TRPV1wt::DTAfl/wt) mice, capsaicin stimulation decreased NK cells expression of GM-CSF. Capsaicin had no impact on NK cell activation when co-cultured with DRG neurons harvested from nociceptor ablated (TRPV1cre::DTAfl/wt) mice (Figure 1A,B). It is worth noting that TRPV1cre mediated cell ablation using diphtheria toxin A (DTA) will eliminate all TRPV1+ neurons, peptidergic and non-peptidergic alike (MrgD+ cells)13,14.
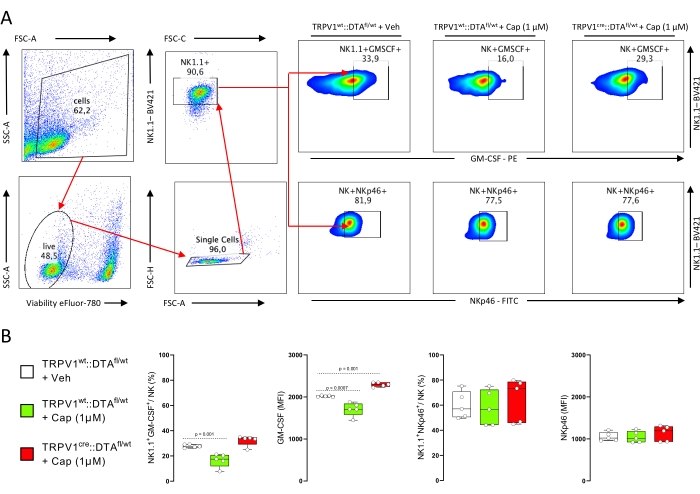
Figure 1: TRPV1+ neurons control NK cell activation. NK cells were magnetically purified from littermate control (TRPV1wt::DTAfl/wt) mice splenocytes and stimulated (48 h) with IL-2 and IL-15. The NK cells were then cultured alone or co-cultured with DRG neurons harvested from nociceptor neuron intact (littermate control; TRPV1wt::DTAfl/wt) or ablated (TRPV1cre::DTAfl/wt) mice. The cells were then exposed to the TRPV1 agonist capsaicin (1 µM) or its vehicle. After 24 h of co-culture, the NK cells were FACS-purified and their activation levels were immunophenotyped using flow cytometry (A). When cultured alone, NK cells expressed basal levels of GM-CSF and NKp46. GM-CSF expression decreased when NK cells were co-cultured with DRG neurons harvested from nociceptor intact (TRPV1wt::DTAfl/wt) mice and stimulated with capsaicin (A,B). The level of NKp46 were unaffected (A,B). Capsaicin had no impact on NK cell activation when co-cultured with DRG neurons harvested from nociceptor ablated (TRPV1cre::DTAfl/wt) mice (A,B). Representative FACS plot are shown (A). Data are presented as mean ± S.E.M (B). The experiment was repeated three times and similar conclusions were obtained. Number of animals tested is shown; n = 5 biological replicates/group (B). P values are shown in the figure and determined by one-way ANOVA post-hoc Bonferroni (B). Abbreviations: TRPV1 = transient receptor potential cation channel subfamily V member 1; NK = natural killer; DTA = diphtheria toxin A; DRG = dorsal root ganglion; FACS = fluorescence-activated cell sorting; GM-CSF = granulocyte-macrophage colony-stimulating factor; Veh = vehicle; Cap = capsaicin. Please click here to view a larger version of this figure.
| Solution | Composition | ||
| Collagenase/Dispase (C/D) | Sterile PBS supplemented with collagenase (1 mg/mL) and Dispase II (2.4 U/mL) | ||
| DMEM | Sterile DMEM supplemented with FBS (10%), penicillin (100 I.U.) and streptomycin (100 µg/mL) | ||
| Supplemented RPMI 1640 | Sterile RMPI 1640 supplemented with penicillin (100 I.U.), streptomycin (100 µg/mL), fetal bovine serum (10%), L-glutamine (300 ng/mL), mouse recombinant IL-2 (200 U/mL), and mouse recombinant IL-15 (10 ng/mL) | ||
| Neurobasal | Sterile Neurobasal supplemented with penicillin (100 I.U.), streptomycin (100 µg/mL), L-glutamine (200 μM), B-27 (2%), NGF (50 ng/mL), and GDNF (2 ng/mL) | ||
| Neurobasal MX | Sterile Neurobasal supplemented with penicillin (100 I.U.), streptomycin (100 µg/mL), L-glutamine (200 μM), B-27 (2%), NGF (50 ng/mL), GDNF (2 ng/mL), mouse recombinant IL-2 (200 U/mL), and mouse recombinant IL-15 (10 ng/mL) | ||
| FACS sorting buffer | Sterile PBS supplemented with FBS (2%) and EDTA (1 mM) | ||
Table 1: List of solutions and media used in this protocol. Abbreviations: NGF = nerve growth factor; GDNF = glial cell-derived neurotrophic factor; FBS = fetal bovine serum; PBS = phosphate-buffered saline.
Discussion
Davies et al.11 found that injured neurons upregulate RAE1. Via cell-cell contact, NKG2D-expressing NK cells were then able to identify and eliminate RAE1+ neurons, which in turn limit chronic pain11. Given that NK cells also express various neuropeptide receptors, and that those neuropeptides are known for their immunomodulatory capabilities, it appears increasingly important to study the interaction between NK cells and nociceptor neurons in vitro. Here we devised a simple co-culture protocol that allows investigators to study how neurons control NK cell function, and reciprocally, how NK cells could modulate the function of DRG neurons. Flow cytometry was used to immunophenotype how neurons modulate NK cell activation. Alternatively, investigators could use qPCR or RNA-sequencing to measure transcript changes in FACS-purified cells or analyze the secretion of secreted factors, such as cytokines or neuropeptides, using ELISAs. Raw data are deposited and freely available on www.talbotlab.com.
Here we found that TRPV1+ nociceptor neurons, secondary to the action of capsaicin, block NK cell activation (GM-CSF expression). While capsaicin used at a high concentration may impair NK cell functions14, we ruled out this possibility experimentally as NK cell activity was not impacted by capsaicin (1 µM) exposure in the absence of peptidergic neurons (TRPV1cre::DTAfl/wt). As capsaicin leads to calcium influx and their subsequent release in neuropeptides, these data suggest that NK cell activation is therefore likely secondary to the action of soluble factors being released by neurons. Therefore, such a co-culture approach was useful to study how the release of neuropeptides may modulate NK cell function. In addition to soluble factors, the local cell-cell interactions between the two cell types are also likely to modulate their functions, something that should be tested experimentally (e.g., comparing the impact of conditioned medium to the one of co-culture).
Overall, such an interplay suggests that within the tissue microenvironment, NK cell function is likely tuned by nociceptor neuron terminals. These data stress the importance of assessing inflammatory responses holistically by simultaneously studying the role of neurons and innate immune cells (i.e., NK cells). For instance, NK cell-neuron crosstalk likely plays an important biological role in contexts spanning from nerve injury, tissue injury, bacterial or viral infection to malignancies. Future work, including the use of this co-culture method, is needed to assess the impact of neuron-NK cell interplay in health and disease.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was supported by The New Frontiers in Research Fund (NFRFE201901326), the Canadian Institutes of Health Research (162211, 461274, 461275), the Canadian Foundation for Innovation (37439), Canada Research Chair program (950-231859), Natural Sciences and Engineering Research Council of Canada (RGPIN-2019-06824), and the Fonds de Recherche du Québec Nature et technologies (253380).
Materials
| Anti-mouse CD16/32 | Jackson Laboratory | Cat no: 017769 | |
| B-27 | Jackson Laboratory | Cat no: 009669 | |
| Bovine Serum Albumin (BSA) culture grade | World Precision Instruments | Cat no: 504167 | |
| BV421 anti-mouse NK-1.1 | Fisher Scientific | Cat no: 12430112 | |
| Cell strainer (50 μm) | Fisher Scientific | Cat no: A3160702 | |
| Collagenase IV | Fisher Scientific | Cat no: 15140148 | |
| Diphteria toxinfl/fl | Fisher Scientific | Cat no: SH3057402 | |
| Dispase II | Fisher Scientific | Cat no: 13-678-20B | |
| Dulbecco's Modified Eagle Medium (DMEM) | Fisher Scientific | Cat no: 07-200-95 | |
| EasySep Mouse NK Cell Isolation Kit | Sigma | Cat no: CLS2595 | |
| Ethylenediaminetetraacetic acid (EDTA) | Sigma | Cat no: C0130 | |
| FACSAria III | Sigma | Cat no: 04942078001 | |
| Fetal bovine serum (FBS) | Sigma | Cat no: 806552 | |
| FITC anti-mouse NKp46 | Sigma | Cat no: L2020 | |
| Flat bottom 96-well plate | Sigma | Cat no: 03690 | |
| Glass Pasteur pipette | Sigma | Cat no: 470236-274 | |
| Glial cell line-derived neurotrophic factor (GDNF) | VWR | Cat no: 02-0131 | |
| Laminin | Cedarlane | Cat no: 03-50/31 | |
| L-Glutamine | Gibco | Cat no: A14867-01 | |
| Mouse recombinant IL-15 | Gibco | Cat no: 22400-089 | |
| Mouse recombinant IL-2 | Gibco | Cat no: 21103-049 | |
| Nerve Growth Factor (NGF) | Life Technologies | Cat no: 13257-019 | |
| Neurobasal media | PeproTech | Cat no: 450-51-10 | |
| PE anti-mouse GM-CSF | PeproTech | Cat no: 212-12 | |
| Penicillin and Streptomycin | PeproTech | Cat no: 210-15 | |
| Pestles | Stem Cell Technology | Cat no: 19855 | |
| Phosphate Buffered Saline (PBS) | Biolegend | Cat no: 108732 | Clone PK136 |
| RPMI 1640 media | Biolegend | Cat no: 137606 | Clone 29A1.4 |
| TRPV1Cre | Biolegend | Cat no: 505406 | Clone MP1-22E9 |
| Tweezers and dissection tools. | Biolegend | Cat no: 65-0865-14 | |
| U-Shaped-bottom 96-well plate | Biolegend | Cat no: 101319 | |
| Viability Dye eFlour-780 | Becton Dickinson |
Referências
- Berta, T., Qadri, Y., Tan, P. H., Ji, R. R. Targeting dorsal root ganglia and primary sensory neurons for the treatment of chronic pain. Expert Opinion on Therapeutic Targets. 21 (7), 695-703 (2017).
- Baral, P., et al. Nociceptor sensory neurons suppress neutrophil and gammadelta T cell responses in bacterial lung infections and lethal pneumonia. Nature Medicine. 24, 417-426 (2018).
- Binshtok, A. M., et al. Nociceptors are interleukin-1beta sensors. Journal of Neuroscience. 28 (52), 14062-14073 (2008).
- Chesne, J., Cardoso, V., Veiga-Fernandes, H. Neuro-immune regulation of mucosal physiology. Mucosal Immunology. 12 (1), 10-20 (2019).
- Samad, T. A., et al. Interleukin-1beta-mediated induction of Cox-2 in the CNS contributes to inflammatory pain hypersensitivity. Nature. 410 (6827), 471-475 (2001).
- Godinho-Silva, C., et al. Light-entrained and brain-tuned circadian circuits regulate ILC3s and gut homeostasis. Nature. 574, 254-258 (2019).
- Talbot, J., et al. Feeding-dependent VIP neuron-ILC3 circuit regulates the intestinal barrier. Nature. 579, 575-580 (2020).
- Raulet, D. H., Gasser, S., Gowen, B. G., Deng, W., Jung, H. Regulation of ligands for the NKG2D activating receptor. Annual Review of Immunology. 31, 413-441 (2013).
- Vivier, E., et al. Innate or adaptive immunity? The example of natural killer cells. Science. 331, 44-49 (2011).
- Davies, A. J., et al. Natural Killer Cells Degenerate Intact Sensory Afferents following Nerve Injury. Cell. 176, 716-728 (2019).
- Perner, C., Sokol, C. L. Protocol for dissection and culture of murine dorsal root ganglia neurons to study neuropeptide release. STAR Protocols. 2, 100333 (2021).
- Goswami, S. C., et al. Molecular signatures of mouse TRPV1-lineage neurons revealed by RNA-Seq transcriptome analysis. Journal of Pain. 15, 1338-1359 (2014).
- Mishra, S. K., Tisel, S. M., Orestes, P., Bhangoo, S. K., Hoon, M. A. TRPV1-lineage neurons are required for thermal sensation. EMBO J. 30, 582-593 (2011).
- Kim, H. S., et al. Attenuation of natural killer cell functions by capsaicin through a direct and TRPV1-independent mechanism. Carcinogenesis. 35, 1652-1660 (2014).

