In Vitro Assay of Plasmodium-Infected Red Blood Cell Killing by Cytotoxic Lymphocytes
Summary
Here we describe a new method to help elucidate the mechanisms of cellular immunity to Plasmodium during the blood stage of infection. This is an in vitro assay that measures infected red blood cell killing by cytotoxic lymphocytes.
Abstract
Malaria is a major public health concern, presenting more than 200 million cases per year worldwide. Despite years of scientific efforts, protective immunity to malaria is still poorly understood, mainly due to methodological limitations of long-term Plasmodium culture, especially for Plasmodium vivax. Most studies have focused on adaptive immunity protection against malaria by antibodies, which play a key role in controlling malaria. However, the sterile protection induced by attenuated Plasmodium sporozoites vaccines is related to cellular response, mainly to cytotoxic T lymphocytes, such as CD8+ and gamma delta T cells (γδ T). Hence, new methodologies must be developed to better comprehend the functions of the cellular immune response and thus support future therapy and vaccine development. To find a new strategy to analyze this cell-mediated immunity to Plasmodium blood-stage infection, our group established an in vitro assay that measures infected red blood cell (iRBC) killing by cytotoxic lymphocytes. This assay can be used to study cellular immune response mechanisms against different Plasmodium spp. in the blood stage. Innate and adaptative cytotoxic immune cells can directly eliminate iRBCs and the intracellular parasite in an effector:target mechanism. Target iRBCs are labeled to evaluate cell viability, and cocultured with effector cells (CD8+ T, γδ T, NK cells, etc.). The lysis percentage is calculated based on tested conditions, compared to a spontaneous lysis control in a flow cytometry-based assay. Ultimately, this killing assay methodology is a major advance in understanding cell-mediated immunity to blood-stage malaria, helping uncover new potential therapeutic targets and accelerate the development of malaria vaccines.
Introduction
Malaria remains a global health crisis, with more than 240 million cases and 627,000 malaria-related deaths reported in 20201. There are currently five parasitic species that can cause malaria in humans, out of which Plasmodium falciparum and Plasmodium vivax are the two most prevalent species. During Plasmodium infection, the liver or pre-erythrocytic stage is asymptomatic, and symptoms only occur during the parasite's asexual cycle in the erythrocytic stage. At this infection stage, thousands of merozoites derived from the liver stage are released into the bloodstream and infect red blood cells (RBCs). In the RBC, the parasites differentiate into trophozoites and schizonts by schizogony, until schizonts rupture the erythrocyte, releasing newly formed merozoites, repeating this blood cycle. Repeated cycles of invasion, replication, and merozoite release result in an exponential growth of the parasite population and ultimately trigger disease symptoms2.
An important challenge in studying the immune response to malaria is that the Plasmodium spp. that infects humans does not infect laboratory animal models. Thus, Plasmodium-infected patient samples must be collected fresh and immediately processed and analyzed. However, in malaria-endemic areas, resources to access immunological and molecular mechanisms are limited. Due to these limitations, rodents are widely used as experimental models to investigate the immune response against Plasmodium infection. While P. berghei and P. chabaudi are often used as surrogates for P. falciparum infection, the non-lethal strain of P. yoelii 17XNL also has many features in common with P. vivax, such as reticulocyte restricted infection3,4. The development of Plasmodium in vitro assays, that can be used for human or animal model-derived samples, is valuable in gaining a better understanding of the pathogenesis of malaria and comparing the immunological response elicited by different species of the parasite.
Protective antimalarial immunity is not completely understood either at the pre-erythrocytic stage nor the blood stage. It is known that exposure to repeated infections results in partial acquired immunity, but sterile immunity is rarely developed5. For decades, anti-Plasmodium protective immunity was mainly associated with the induction of neutralizing or opsonizing antibodies that prevent parasite invasion of host cells or lead to phagocytosis by antigen-presenting cells, respectively6. As a result, most efforts to produce antimalarial vaccines thus far have relied on inducing protective and long-lasting antibodies7,8. However, the sterile protection induced by vaccination with an attenuated sporozoite directly correlates with the activation and expansion of cytotoxic T lymphocytes8,9.
Recently, some studies of freshly isolated patient samples and in vitro cultures have demonstrated that innate or adaptative cytotoxic immune cells like CD8+ T10, γδ T11, and NK cells12 can directly eliminate Plasmodium-infected RBCs and its intracellular parasite in an effector:target ratio manner. These seminal findings defined an entirely new immune effector mechanism in the context of malaria. To dissect this novel antimalarial immunity, it is essential to explore cytotoxic effector mechanisms of killer cells against infected-RBCs (iRBCs) in natural infection or vaccination.
Here we present an in vitro assay that measures the cytotoxic activity of lymphocytes against malaria in the blood stage. This assay can then help elucidate the mechanisms of the cellular immune response against the Plasmodium erythrocyte stage. The target cells, iRBCs, are labeled with carboxyfluorescein succinimidyl ester (CFSE) to evaluate cell viability, and are then cocultured with effector cells like cytotoxic lymphocytes (CTL). This coculture is then evaluated by flow cytometry, using fluorescent markers for specific cell types. Finally, the percentage of iRBC lysis by CTL is calculated by dividing the experimental condition by the spontaneous rupture of RBCs and spontaneous lysis control, which occurs during incubation without the effector cell. Overall, this killing assay methodology can contribute to a better understanding of cell-mediated malaria immunity.
Protocol
All procedures were conducted following the policies of the Oswaldo Cruz Foundation and the National Ethical Council (CAAE: 59902816.7.0000.5091). The human protocols were developed in collaboration with the clinical research group from the Research Center for Tropical Medicine of Rondônia (CEPEM), which was in charge of enrolling patients in the study. Informed consent was obtained from all the patients.
For the animal study, procedures were performed following the principles of conduct of the Brazilian Practice Guide for the Care and Use of Animals for Scientific and Didactic Purposes of the National Council for the Control of Animal Experimentation (CONCEA). The protocols were approved by the Fiocruz Animal Experimentation Council (CEUA protocol LW15/20-2).
1. Collection of human blood samples and PBMC isolation
- Collect blood of Plasmodium-infected patients in a 10 mL evacuated blood collection tube with sodium heparin. As malaria patients display lymphopenia, there is a range of 5-9 x 106 peripheral blood mononuclear cells (PBMCs) in a 10 mL blood sample. Since CD8+ T cells or γδ T cells make up ~10% of PBMCs, preferably collect 50-100 mL of blood/patient.
NOTE: A minimum of 1 x 105 effector cells/ condition should be considered. - Calculate the percentage of iRBCs by blood smear as described below.
- Add 5 μL of total blood to a clear slide and prepare a blood smear. Perform Romanowsky-type panoptic fast stain or May-Günwald-Giemsa stain. Here, the panoptic fast stain kit is used, which is composed of three reagents: reagent A (fixation), reagent B (cytoplasmic stain), and reagent C (nuclear and cytoplasmic differential stain).
- Slowly dip the slide in solution A 10x, then 4x in solution B, and finally 10x in solution C. Drain out any excess reagent from the slides between solutions. After dipping in solution C, rinse the slide in running tap water and let it dry.
- Under an upright light microscope with a 100x oil immersion objective, count 1000 RBCs in sequential squares and calculate the percentage of parasitemia using the following equation:
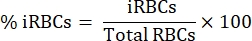
- Dilute 15 mL of blood in a 1:1 proportion in sterile phosphate-buffered saline (PBS).
- Add 15 mL of lymphocyte separation medium (density of 1.077g/mL) to a 50 mL tube. Carefully layer the 30 mL of diluted blood sample onto the centrifugation medium solution. When layering the sample, do not let the blood sample and lymphocyte separation medium mix.
- Centrifuge the tubes at 400 x g for 40 min at 22 °C, with low acceleration and no break setting.
- Draw off the upper layer containing plasma using a sterile pipette, leaving the mononuclear cell layer undisturbed. Transfer the layer of mononuclear cells (PBMCs) to a sterile tube using a sterile pipette.
- Do not discard the tube containing the blood pellet as it will be used later for infected RBC isolation. From this step, be sure to keep the RBCs at room temperature (RT). Never let them cool down.
- Wash the cells twice by adding PBS and centrifuge at 350 x g for 10 min, with break setting. Resuspend the cell pellet in 5 mL of RPMI medium supplemented with penicillin/streptomycin and 10% fetal bovine serum (FBS; complete medium).
- Count the PBMCs in the presence of trypan blue solution to check cell viability, using a hemacytometer (Neubauer chamber) or automated cell counter. Do not count the blue-stained cells as these represent dying cells, which absorb trypan blue.
- Adjust the cell concentration to 107 cells/mL using complete medium. Purify the desired cytotoxic lymphocyte populations (CD8+ T cells, NK cells, γδ T cells) using magnetic beads isolation as per reagent manufacturer protocol.
2. Human RBC isolation
NOTE: For human-infected RBC isolation, it is recommended to start with blood samples that have a minimum of 2% parasitemia, preferentially with more in the trophozoite/early schizont parasite stage.
- Prepare the PERCOLL (from hereafter referred to as density gradient separation medium) at the recommended concentration as described below.
- Add 90 mL of 100% density gradient separation medium and 10 mL of 10x PBS to obtain 90% isotonic density gradient separation medium.
- For P. vivax-infected reticulocyte separation, prepare45% density gradient separation media. Add 50 mL of 90% isotonic density gradient separation medium and 50 mL of 1x PBS to obtain 45% density gradient separation medium.
- For P. falciparum-infected erythrocyte separation, prepare 65% density gradient separation media. Add 72 mL of 90% isotonic density gradient separation medium and 28 mL of 1x PBS to obtain 65% density gradient separation medium.
- For uninfected reticulocyte separation, prepare 70% density gradient separation media. Add 78 mL of 90% isotonic density gradient separation medium and 22 mL of 1x PBS to obtain 70% density gradient separation medium.
- Warm the density gradient separation medium to 37 °C in a water bath.
- After removal of the PBMC layer, carefully remove as much of the top layer of centrifugation medium as possible without touching the cells. With a glass Pasteur pipette, carefully remove the upper neutrophil layer without disturbing the RBC pellet and estimate pellet volume. Add 4x the RBC pellet volume of RT complete medium and resuspend.
- In a second 50 mL conical tube, add 5x the pellet volume of 45%, 65%, or 70% density gradient separation medium depending on the cell type to isolate. Layer the RBC suspension on top of the density gradient separation medium layer carefully.
- Spin for 15 min at 850 x g, with low acceleration and no break setting. Collect the cloudy red/brown layer lying in between the supernatant and density gradient separation medium with a 5 mL pipette. Transfer to a sterile 15 mL tube.
- Wash the cells by adding RT complete medium up to 15 mL and spin down for 10 min at 860 x g. Repeat the washing by adding 10 mL of RT complete medium and spin down. Discard the supernatant and prepare a blood smear from the pellet to check for iRBC enrichment, following step 1.2.
- Resuspend the pellet in 1 mL of RT complete medium and let sit at room temperature while counting the cells. Add 10 μL of cell suspension in a hemocytometer (Neubauer chamber) and count the RBCs in the central area subdivided into 25 medium squares. Adjust cell concentration to 1 x 107 iRBCs/mL in RT complete media.
3. Experimental malaria infection in mouse
- Thaw an aliquot of cryopreserved P. yoelii 17XNL:PyGFP (MRA-817), a GFP-expressing strain obtained from MR4/ATCC, and inject 100 μL intraperitoneally (i.p.) in an 8-week-old female C57BL/6 mouse.
- Follow the parasitemia burden every 3 days by tail vein lancing blood collection.
- Puncture the vessel with the needle bevel up, entering the vein at a shallow angle beginning at the distal end of the tail.
- Collect the blood sample with a pipette or capillary tube up to 5 or 10 mL, then apply manual pressure to stop the bleeding.
- Prepare a blood smear (as described in step 1.2) until parasitemia reaches 10%-15% of infected RBCs.
- Collect 10 μL of blood by tail vein lancing and dilute to 100 μL of PBS to i.p. inject into the second donor mouse.
NOTE: Cryopreserved parasites should be thawed and passed in mice twice before being used for experimental infections. - When the second passage reaches 15% of parasitemia, collect five drops of blood by the tail clipping method and dilute in 1 mL of PBS. Prepare an infection solution by adjusting the concentration to 1 x 106 iRBCs/mL in sterile PBS and inject i.p. 100 μL (1 x 105 iRBCs) of the solution into each mouse required for the experiment.
- Monitor parasitemia every 2-3 days until it reaches ~30% iRBCs, which occurs approximately 12 days post-infection. When the mice reach the desired parasitemia, collect blood by cardiac puncture as described below.
- Aspirate 100 µL of heparin solution (30 U/mL) into a 1 mL syringe with a 26 G needle.
- Anesthetize the mouse by inhalation with 5% isoflurane and confirm the absence of reflexes. Place the mouse on its side and perpendicularly insert the needle just below the elbow, through the ribs, and into the heart. Pull out the syringe plunger slowly and rotate the needle until 0.5-1 mL of blood is obtained.
- Perform humane euthanasia by cervical dislocation under anesthesia. Aseptically clean the left side of the mouse with 70% ethanol. With surgical scissors, make a cut on the left side of the mouse passing through the skin and peritoneum. Locate the spleen and remove it.
- Place the mouse spleen in a Petri dish with 5 mL of complete medium to purify the desired effector cell population (e.g., CD8+ T cells).
4. Obtaining fresh whole mouse splenocytes
- In the Petri dish, carefully cut the spleen into small pieces using scissors or a razor.
- Place a 100 µM cell strainer over a 50 mL conical tube and transfer the excised spleen into the cell strainer with a pipette. Mash the spleen through the strainer with a syringe plunger.
- Wash the cells through the strainer with 10 mL of complete media. Centrifuge the cells at 300 x g for 10 min at 4 °C, then discard the supernatant.
- Resuspend the cell pellet in 2 mL of cold 1x RBC lysis buffer. Incubate the suspension for 5 min on ice.
- Wash the cell suspension with 10 mL of 4 °C complete media. Repeat the wash step three times and remove any cell clots between washes for spleens from Plasmodium-infected mice.
- Centrifuge the cells at 400 x g for 5 min at 4 °C, then discard the supernatant.
NOTE: Plasmodium-infected spleens are enlarged and filled with phagocytic cells containing degraded hemoglobin and hemozoin. - To avoid any issues during the magnetic bead isolation of effector cells, use the following steps to remove hemozoin-enriched phagocytic cells and hemozoin. Resuspend the splenic cells with MACS buffer and adjust concentration to 1 x 108 cells/mL. Place an LS or LD column in the magnetic field. Prepare the column by rinsing with 3 mL of MACS buffer.
- Apply cell suspension onto the column. Wash the column three times with 3 mL of MACS buffer. Collect flow-through containing the splenic cells.
- Count the splenocytes in a trypan blue solution and check cell viability in a hemacytometer (Neubauer chamber) or automated cell counter. Adjust cell concentration to 1 x 107 cells/mL in RT complete media.
5. Cytotoxic effector cell purification (CD8a+ T cell negative selection)
NOTE: There are many positive and negative selection reagents that can purify cytotoxic effector cells (CD8+ T, γδ T, NK, iNKT, MAIT cells). In this protocol, we use a negative selection of splenic CD8a+ T cells and follow manufacturer instructions.
- Centrifuge all splenocytes at 400 x g for 10 min at 4 °C and discard the supernatant. Resuspend the cell pellet in 40 µL of MACS buffer.
- Add 10 µL of a biotin-antibody cocktail. Mix well and incubate for 5 min on ice.
- Add 30 µL of MACS buffer. Add 20 µL of anti-biotin micro-beads. Mix well and incubate for 10 min on ice.
- Add 400 µL of MACS buffer and proceed to magnetic cell separation. Place the MACS LS column in the magnetic field support. Prepare the column by rinsing with 3 mL of MACS buffer.
- Apply cell suspension onto the column. Wash the column three times with 3 mL of MACS buffer. Collect the flow-through containing all unlabeled cells, which are the enriched CD8a+ T cells.
- Count the CD8a+ T cells in a trypan blue solution and check cell viability in a hemacytometer (Neubauer chamber) or automated cell counter. Adjust the cell concentration to 1 x 107 cells/mL in RT complete media.
6. P. yoelii infected RBC isolation
- Centrifuge the collected blood in a 1.5 mL tube at 850 x g for 3 min. Discard the serum and resuspend the blood in 1 mL of 1x RPMI without FBS.
- Place the LS column in the magnetic field support and rinse with 3 mL of 1x RPMI. Pass the RBC suspension through the column. To isolate more iRBCs, reapply the flowthrough (3 mL of RPMI and 1 mL of diluted blood) into the column.
- Wash twice with 5 mL of 1x RPMI. Perform washing steps by adding buffer aliquots as soon as the column reservoir is empty. Do not let any columns dry up.
- Add 5 mL of RPMI, remove the column, and purge the iRBCs into a new 15 mL tube. Count the iRBCs and adjust the concentration to 1 x 107 RBCs/mL.
7. CFSE labeling of RBCs and preparation for flow cytometry
NOTE: Start the RBC labeling protocol with twice the number of cells that will be used for the experiment, since ~50% of the cells are typically lost in the washing steps following the CFSE labeling step. To establish the autofluorescence of the RBCs/iRBCs, include a control sample of unlabeled cells.
- Dilute CFSE to a final concentration of 10 mM in 1x RPMI without FBS. Wash the cells once with 1x RPMI without FBS in a 15 mL tube.
- Resuspend the RBCs in 500 μL of 1x RPMI without FBS and add 500 μL of the diluted CFSE. Incubate for 8 min at RT, protected from light.
- Wash three times by adding 14 mL of complete medium (10% FBS RPMI), followed by centrifugation at 850 x g for 10 min. Resuspend the cells in complete medium to a concentration of 1-5 x 106/ mL. Incubate for 1 h at RT.
8. Cytotoxic lymphocyte/RBC coculture and preparation for flow cytometry
NOTE: If the involvement of specific receptors or molecules will be measured, incubate the specific blocking and isotype control antibodies (10 mg/mL) with the effector cells for 30 min before coculture.
- Plate the cells in a 96-well round-bottom plate. Add the purified lymphocytes and CFSE-labeled iRBCs in the desired effector-target cell ratio to a final volume of 200 μL and homogenize. Prepare each condition in triplicate.
NOTE: We suggest adjusting the iRBC concentration to 1-5 x 104 cells/mL and choosing the ratio based on the purified cell number (e.g., 0.5:1, 2.5:1, and 5:1). - Include the spontaneous lysis control, which is the target iRBCs without the effector, to evaluate any spontaneous lysis, as this will be used to represent a 100% cell viability condition.
- Spin down the plate for 1 min at 360 x g to maximize cell contact. Incubate at 37 °C and 5% CO2 for 4 h.
NOTE: Hypoxia conditions (low O2), systematically used for P. falciparum culture, should not be used since the effector cells do not survive in this environment. In contrast, Plasmodium parasites are viable in ambient air in 5% CO2 for up to 12 h. - Spin for 5 min at 850 x g and flip the plate to remove the supernatant.
NOTE: If desired, the supernatant can be used to measure any soluble factors released in the coculture. - Label the RBCs with anti-mouse Ter119 1:200 (or anti-human CD235 1:100 for human samples) and CD8+ T cells with anti-CD8 1:200 or anti-CD3 1:200 antibodies (anti-mouse or human) for 30 min at 4 °C in 1x PBS containing 3% FBS (FACS buffer).
NOTE: The antibody fluorophore should be selected based on the cell tracer/cell proliferation reagent (e.g., CFSE-labelled iRBC, APC-Cy7 anti-Ter119, and PerCP-Cy5.5 anti-CD8a). - Wash the cells with FACS buffer and spin down for 5 min at 850 x g. Transfer the samples to FACS tubes and add 30 μL of counting beads to individual tubes. Homogenize the count beads by vortexing for 30 s.
9. Flow cytometry
- Analyze samples using a 405/488/561/640 laser instrument.
- For human cells labeled with CFSE (FITC), PE anti-human γδ TCR, and PE-Cy7 anti-human CD235a antibody, use 530/30 (FITC), 575/25 (PE), and 780/60 (PE-Cy7) filters in a three-laser (Blue, Red, Yellow-Green) configuration cytometer.
- For mouse cells labeled with CFSE (FITC), PerCP-Cy5.5 anti-mouse CD8a, and APC-Cy7 anti-mouse Ter119, use 530/30 (FITC), 695/40 (PerCP-Cy5.5), and 780/60 (APC-Cy7) filters in the three-laser cytometer setup.
- Choose the counting beads population as the stopping gate to acquire a minimum of 20,000 events in the stop gate, which should be identical for all conditions. Perform analysis and compensation using appropriate flow cytometry acquisition/analysis software.
- Set the gating strategy as described below.
- Select single cells (singlets), excluding debris using FSC peak height (H) to area (A) ratio. Select RBCs and exclude the lymphocytes from analysis based on fluorescence of the RBC marker (Ter119 or CD235a) and the lymphocyte-specific antibody (CD8a).
- In the previous RBC gating, select viable RBCs based on CFSE positive staining. Analyze the data with flow cytometer data analysis software.
10. Calculation and statistics
- To calculate the percentage of iRBC lysis, follow the gating strategy described in step 9. The percentage of viable RBCs is the frequency of CFSE-positive cells inside the RBC gate based on Ter119 (mouse) or CD235a (human) positive fluorescence.
- Use the spontaneous lysis control, RBCs without effector cells, condition as a control to estimate RBC spontaneous rupture. This condition will be considered RBC lysis (100% viability). Use the following formula to calculate the percentage of RBC lysis in each tested condition:
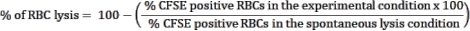
NOTE: During the culture incubation, some infected RBCs may spontaneously lyse or be ruptured by the parasite. Use the CFSE-positive RBC frequency of the spontaneous lysis condition, which contains no effector cells, as the baseline for lymphocyte cytotoxicity. - Calculate the average of triplicates for each condition and determine statistical significance using two-way ANOVA at multiple comparisons.
Representative Results
Here, the applied methodology for isolation of CFSE-labeled Plasmodium-infected RBCs in a coculture assay with cytotoxic lymphocytes is described. First, we provide a schematic representation of how to perform the protocol, employing human samples infected with P. vivax (Figure 1). Then, an illustrated flowchart on how to proceed with the protocol in a malaria experimental model using a C57BL/6 mouse infected with P. yoelii (Figure 2). In Figure 3, the expected results (before and after) for step 6 of the protocol (enrichment of P. yoelii-infected RBCs using magnetic columns) are shown. Finally, Figure 4 shows a representative flow cytometry analysis, detailing the gating strategy needed to assess the percentage of RBC lysis, as described in step 9. Therefore, this section highlights the different techniques used here, describing the entire process of acquisition, assembly, and data analysis to assess iRBC killing by cytotoxic lymphocytes.
Human Plasmodium-infected RBCs lysis by cytotoxic cells
A schematic representation of the protocol to evaluate the killing of human Plasmodium-infected RBCs by cytotoxic cells is shown in Figure 1. To measure cell lysis, PBMC from Plasmodium-infected patients was purified using the separation medium, followed by magnetic purification of the cytotoxic lymphocyte (CTL) population of interest (e.g., CD8+ T, γδ T, NK, iNKT, MAIT cells, etc.). The RBCs' pellet remaining from the PBMC isolation was used to enrich Plasmodium-infected RBCs using density gradient separation medium (65% P. falciparum; 45% P. vivax). iRBCs were labeled with CFSE and incubated with or without lymphocytes for 4 h at 37 °C, 5% CO2. After the coculture period, all RBCs (infected or not) were labeled with anti-human CD235a antibody (for the RBCs) and CTL-specific antibodies (e.g., anti-CD8, anti- γδTCR, etc.) before flow cytometry analysis. The acquisition and analysis of human samples were similar to those demonstrated for mouse samples in Figure 4.
P. yoelii-infected RBC lysis by cytotoxic CD8+ T cells
A flow chart of the experimental procedure to evaluate the cytotoxic effector mechanism in the experimental malaria model is shown in Figure 2, along with representative experimental data shown in Figure 3 and Figure 4. C57BL/6 mice were infected with 1 x 105 P. yoelii (Py) iRBCs, and parasitemia was monitored until it reached around 30%. Py-iRBCs were isolated from blood using magnetic separation, as the parasite heme subproduct, hemozoin, is iron-enriched and functions as paramagnetic particles in a magnetic field13. The purity of the enriched sample was analyzed by blood smear in comparison with the pre-column sample (Figure 3). iRBCs were then labeled with the cell tracer reagent CFSE. In parallel, cytotoxic CD8+ T cells were purified from splenocytes using a magnetic negative selection kit. As a control, the same protocol was used for CD8+ T cell purification from uninfected mice. Effector (CD8+ T) and target (CFSE-labelled Py-iRBC) cells were incubated at different effector:target ratios (E:T: 0.5:1, 2.5:1, and 5:1) for 4 h at 37 °C 5% CO2. At the end of the coculture, each condition was labeled with antibodies anti-mouse Ter119 for RBCs and anti-CD8a for cytotoxic cells. The gating strategy and sample results are represented in Figure 4.
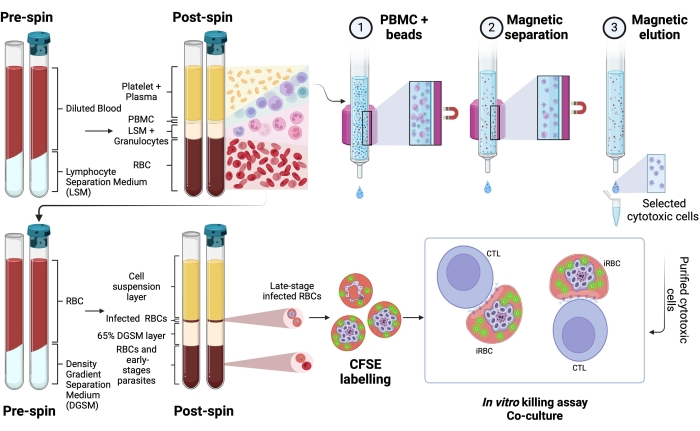
Figure 1. Schematic of the in vitro killing assay. Example of killing assay experiment. Plasmodium-infected blood is collected and processed using a separation gradient medium. After centrifugation, the PBMC layer is collected, and cytotoxic cells (CTLs) were purified using magnetic beads. The RBC layer is once again processed using density gradient separation media. After centrifugation, the Plasmodium-iRBC layer is collected and labeled with CFSE. Thereafter, the purified CTL and iRBCs were cocultured for 4 h at 37 °C, 5% CO2, followed by cell-specific labeling and analysis in a flow cytometer. Please click here to view a larger version of this figure.
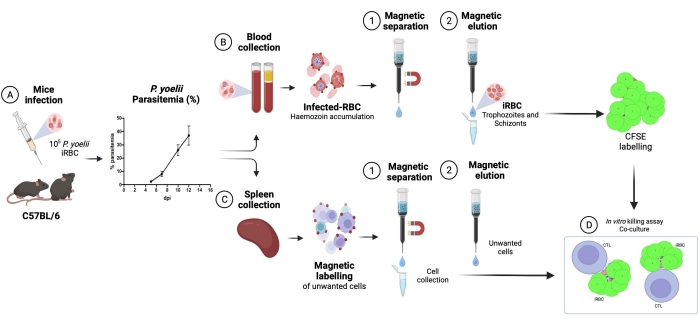
Figure 2. Experimental malaria model for killing assay. First, C57BL/6 mice were infected with 105 PyX17NL-infected red blood cells. Then, parasitemia is monitored by blood smear, until the peak of infection (30%-40% iRBCs), approximately 12 days post-infection (dpi). On the next day, mice were bled and euthanized for spleen collection. The blood was processed, and infected RBCs were enriched by magnetic separation, followed by labeling with CFSE (upper right). The spleen was processed for cytotoxic lymphocytes isolation, by negative selection using magnetic beads. Afterward, the purified cytotoxic lymphocytes and iRBCs were cocultured for 4 h at 37 °C, 5% CO2, followed by cell-specific labeling and analysis in a flow cytometer. Please click here to view a larger version of this figure.
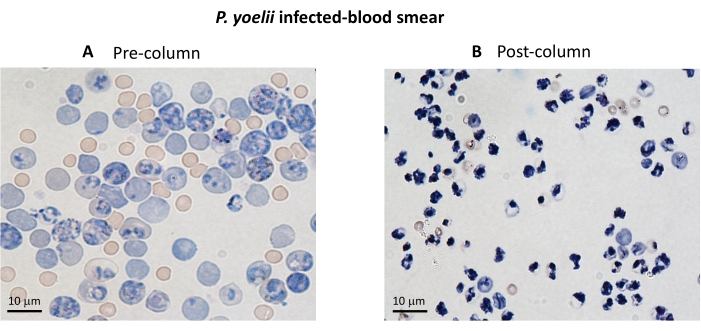
Figure 3. iRBC enrichment using magnetic columns. The iRBCs were purified from P. yoelii infected mice using LS columns. The purification is highlighted by blood smears from blood collected before (A) and after (B) column enrichment. Scale bar = 10 µm. Please click here to view a larger version of this figure.
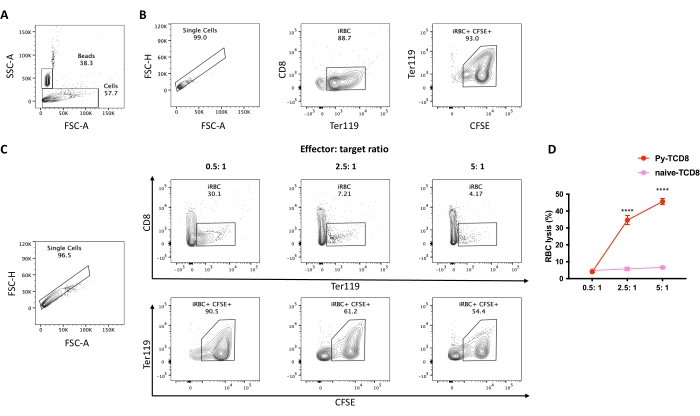
Figure 4. Assessment of cell lysis percentage of P. yoelii-iRBCs by CD8+ lymphocytes. The gating strategy for cell lysis percentage. (A) Set the stopping gate into the counting beads population (~20,000 events). (B) Control spontaneous lysis, iRBCs without lymphocytes. Start the gating strategy by selecting the single cells, excluding debris, based on the FSC peak height to area ratio, followed by the selection of the RBC population in APC-Cy7 Ter119 positive and PerCP-Cy5.5 CD8 negative. Then select live iRBCs as CFSE (FITC) high positive. (C) Example experiment analysis using different effector: target ratios, displaying the percentage of live RBC after the coculture. (D) Graphical representation of the RBC lysis percentage calculated based on the formula described in section 10. The significance comparison between the groups (cytotoxic CD8 or naïve CD8) was assessed by two-way ANOVA with multiple comparisons. P < 0.0001 (****). Please click here to view a larger version of this figure.
Discussion
Here we describe an in vitro assay to measure Plasmodium-infected red blood cell killing by cytotoxic lymphocytes. This assay can help elucidate the mechanisms of cellular protective immunity to the malaria parasite's erythrocytic stage. The major advantage of this methodology is that it provides a quantitative assay of the cell-mediated killing of iRBCs that can be used to address many questions about how immune cells interact with different Plasmodium spp.
Importantly, this method can be used to study P. vivax and other Plasmodium parasites that are not maintained in vitro culture14,15. In addition, this protocol can be adapted to any Plasmodium species and different hosts, such as humans, mice, and non-human primates. Flow cytometry-based methods are widely used to evaluate cytotoxic cell activation, cytokine production, and degranulation10,11,16, but the current method is the only one that explores RBC lysis by direct cell contact with killer cells.
There are also many methods, such as microscopy, polymerase chain reaction (PCR), and flow cytometry, that are used to study malaria by measuring parasitemia, invasion, and antimalarial activity. However, to analyze the cell-mediated cytotoxicity of RBCs, current methods that use Cr51 or hemoglobin release are not sensitive enough to evaluate RBC lysis. LDH release may also be used to evaluate RBC killing by some CTLs like CD8+ T cells, but measuring LDH is not an accurate method as activated innate or innate-like cells may kill each other as a regulatory mechanism.
CFSE is widely used to track cells or explore cell proliferation in flow cytometry or fluorescent microscopy17. Since RBCs do not proliferate18, the CFSE fluorescence intensity should decrease with cell lysis. Flow cytometry is a very accurate technique as it can detect very low levels of specific markers, and so it has been widely used to track parasitemia and invasion in malaria infection19,20,21,22,23. Most importantly, flow cytometry can measure many parameters in a single cell and sort out cells into different populations. By adding quantitative standard reagents (counting beads) to each sample, one can normalize the number of acquired events and obtain absolute cell counts24. This method allows for a reliable quantitative comparison between experimental conditions, which is critical for the currently proposed protocol.
Some critical steps for this protocol include ensuring the sample quality, ensuring proper cell staining, and including controls to normalize calculations. A large amount of fresh blood samples is required to purify infected RBCs. The density gradient separation medium purification should be approached carefully as its concentration is critical to obtaining the desired Plasmodium iRBCs. The blood overlay onto the density gradient separation medium must also be done as slowly as possible, and the ring red-layer harvesting must be done carefully to avoid losing iRBCs. To make the purification process easier, a high parasitemia blood sample should be used as the amount of iRBCs will be higher. It should be noted that in the RBC isolation protocol using magnetic fields, only parasite mature stages (middle-late stage trophozoites or schizonts) that produce significant amounts of hemozoin will be purified. As a result, any ring stage will not be collected, and this may affect the final result.
Cell staining should be performed carefully to avoid loss or lysis of cells. Prolonged storage or fixation should be avoided before staining. It is important to note that the experimental conditions should be performed in triplicate using serial effector:target ratios and appropriate controls, such as an unlabeled, spontaneous lysis condition and a hypothesis control. The addition of cell counting beads is crucial as it provides an easy way to determine the concentration of cells or absolute cell counts for each sample. During flow cytometer acquisition, be sure to set the number of events to be collected in the counting beads gate to normalize the number of cells in each sample after coculture.
Since at least 1 x 105 effector cells are needed for consistent results in a coculture assay, one possible limitation of the described methodology is the limited number of cells after purification, which can be a concern especially when working with rare cell populations. Indeed, this applies to iRBC purification, as it requires having a very high parasitemia that is not so easily obtained, especially when working with human samples in endemic areas.
One protocol improvement may be the coculture incubation time. Although we previously used a 12 h incubation time10,11, we recently observed that 4 h is enough to differentiate the experimental conditions, thus reducing the time for potential spontaneous lysis and improving the reproducibility of the results.
The mechanisms involved in the direct killing of Plasmodium-infected RBCs are gradually being uncovered. Our group was the first to show that CD8+ T cells can recognize and kill P.vivax-infected reticulocytes via HLA-I presentation by the infected cell11. Recently, another study demonstrated that γδ T cells recognize P. falciparum-infected RBCs through phosphoantigen recognition by butyrophylin, a molecule present on the surface of iRBCs10. In both studies, RBC lysis is granulysin and granzyme B dependent, and leads to intracellular parasite killing.
Although many methods have been developed over the years to measure parasitemia, invasion, antimalarial activity, and image cell interaction, none have been capable of analyzing the direct killing of iRBCs by cytotoxic cells in a quantitative flow cytometry-based assay19,20,21. We used an innovative approach to evaluate cell lysis capacity in malaria by CFSE labeling of Plasmodium-infected RBCs and evaluation of iRBC lysis in a specific period of coculture with lymphocytes. Therefore, this in vitro killing assay presents a novel strategy to clarify the mechanisms of cell-mediated immunity to blood-stage malaria, which will help advance the study of new therapeutic targets and the development of malaria vaccines.
Declarações
The authors have nothing to disclose.
Acknowledgements
We thank Dr. Dhelio Pereira and the members of Research Center for Tropical Medicine of Rondônia (CEPEM) for malaria patient enrollment and blood collection and Felicia Ho for helping with manuscript revision. The following reagent was obtained through BEI Resources, NIAID, NIH: Plasmodium yoelii subsp. yoelii, Strain 17XNL:PyGFP, MRA- 817, contributed by Ana Rodriguez. This research was supported by Lemann Brazil Research Fund, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) – 437851/2018-4, fellowships (CJ, GC, CG), and Fundação de Amparo do Estado de Minas Gerais (FAPEMIG) – APQ-00653-16, APQ-02962-18; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) – fellowship (LL).
Materials
| 100 μM cell strainer | Corning | 431752 | |
| 96 Well Round (U) Bottom Plate | Thermo Scientific | 12-565-65 | |
| Anti-human CD235a (Glycophorin A) Antibody | Biolegend | 349114 | Used – APC anti-human CD235, dilution 1:100 |
| Anti-human CD3 Antibody | Biolegend | 317314 | Used – PB anti-human CD3, dilution 1:200 |
| Anti-human CD8 Antibody | Biolegend | 344714 | Used – APC/Cy7 anti-human CD8, dilution 1:200 |
| Anti-human TCR Vδ2 Antibody | Biolegend | 331408 | Used – PE anti-human TCR Vδ2, dilution 1:200 |
| Anti-mouse CD8a Antibody | Biolegend | 100733 | Used- PerCP/Cyanine5.5 anti-mouse CD8a, dilution 1:200 |
| Anti-mouse TER-119/Erythroid Cells Antibody | Biolegend | 116223 | Used – APC/Cyanine7 anti-mouse TER-119, dilution 1:200 |
| CellTrace CFSE Cell Proliferation Kit | Invitrogen | C34554 | |
| Fetal Bovine Serum, qualified | Gibco | 26140079 | |
| Ficoll-Paque Plus | Cytiva | 17144003 | Lymphocyte Separation Medium (LSM) |
| Heparin Sodium Injection, USP | meithel pharma | 71228-400-003 | Used – 2000 USP units/2mL |
| Isoflurane | Piramal critical care | 66794-0013-25 | |
| LS MACS Column | Miltenyi Biotec | 130-042-401 | |
| LSRFortessa Cell Analyzer | BD Bioscience | ||
| Percoll | Cytiva | 17089101 | Density Gradient Separation Medium (DGSM) |
| QuadroMACS Separator | Miltenyi Biotec | 130-090-976 | |
| RPMI 1640 Medium | Gibco | 11875093 | |
| Sodium bicarbonate, powder, BioReagent | Sigma-Aldrich | S5761 | |
| Syringe With Sub-Q needle – 1mL, 26 gauge; | BD | 14-829-10F | |
| Vacutainer Heparin Tube Glass Green 10 ml | BD | 366480 |
Referências
- WHO. Global Technical Strategy for Malaria 2016-2030, 2021 Update. World Health Organization. , (2021).
- Hafalla, J. C., Silvie, O., Matuschewski, K. Cell biology and immunology of malaria. Immunological Reviews. 240 (1), 297-316 (2011).
- Belnoue, E., et al. Vaccination with live Plasmodium yoelii blood stage parasites under chloroquine cover induces cross-stage immunity against malaria liver stage. Journal of Immunology. 181 (12), 8552-8558 (2008).
- Leong, Y. W., Lee, E. Q. H., Rénia, L., Malleret, B. Rodent malaria erythrocyte preference assessment by an ex vivo tropism assay. Frontiers in Cellular and Infection Microbiology. 11, 680136 (2021).
- Ladeia-Andrade, S., Ferreira, M. U., De Carvalho, M. E., Curado, I., Coura, J. R. Age-dependent acquisition of protective immunity to malaria in riverine populations of the amazon basin of Brazil. The American Journal of Tropical Medicine and Hygiene. 80 (3), 452-459 (2009).
- Antonelli, L. R., et al. The immunology of Plasmodium vivax malaria. Immunological Reviews. 293 (1), 163-189 (2020).
- Kazmin, D., et al. Systems analysis of protective immune responses to RTS, S malaria vaccination in humans. Proceedings of the National Academy of Sciences of the United States of America. 114 (9), 2425-2430 (2017).
- Epstein, J. E., et al. Live attenuated malaria vaccine designed to protect through hepatic CD8+ T cell immunity. Science. 334 (6055), 475-480 (2011).
- Draper, S. J., et al. Malaria vaccines: recent advances and new horizons. Cell Host & Microbe. 24 (1), 43-56 (2018).
- Junqueira, C., et al. γδ T cells suppress Plasmodium falciparum blood-stage infection by direct killing and phagocytosis. Nature Immunology. 22 (3), 347-357 (2021).
- Junqueira, C., et al. Cytotoxic CD8+ T cells recognize and kill Plasmodium vivax-infected reticulocytes. Nature Medicine. 24 (9), 1330-1336 (2018).
- Arora, G., et al. NK cells inhibit Plasmodium falciparum growth in red blood cells via antibody-dependent cellular cytotoxicity. eLife. 7, 36806 (2018).
- Paul, F., Roath, S., Melville, D., Warhurst, D. C., Osisanya, J. O. S. Separation of malaria-infected erythrocytes from whole blood: use of a selective high-gradient magnetic separation technique. Lancet. 2 (8237), 70-71 (1981).
- Shaw-Saliba, K., et al. Insights into an optimization of Plasmodium vivax Sal-1 in vitro culture: the aotus primate model. PLOS Neglected Tropical Diseases. 10 (7), 0004870 (2016).
- Mehlotra, R. K., et al. Long-term in vitro culture of Plasmodium vivax isolates from Madagascar maintained in Saimiri boliviensis blood. Malaria Journal. 16 (1), 442 (2017).
- Hojo-Souza, N. S., et al. Contributions of IFN-γ and granulysin to the clearance of Plasmodium yoelii blood stage. PLOS Pathogens. 16 (9), 1008840 (2020).
- Parish, C. R., Glidden, M. H., Quah, B. J. C., Warren, H. S. Use of the intracellular fluorescent dye CFSE to monitor lymphocyte migration and proliferation. Current Protocols in Immunology. 84 (1), 4-9 (2009).
- Migliaccio, A. R. Erythroblast enucleation. Haematologica. 95 (12), 1985 (2010).
- Grimberg, B. T., Erickson, J. J., Sramkoski, R. M., Jacobberger, J. W., Zimmerman, P. A. Monitoring Plasmodium falciparum growth and development by UV flow cytometry using an optimized Hoechst-thiazole orange staining strategy. Cytometry Part A:The Journal of the International Society for Analytical Cytology. 73 (6), 546-554 (2008).
- Pattanapanyasat, K., et al. Culture of malaria parasites in two different red blood cell populations using biotin and flow cytometry. Cytometry:The Journal of the International Society for Analytical Cytology. 25 (3), 287-294 (1996).
- Bianco, A. E., Battye, F. L., Brown, G. V. Plasmodium falciparum: rapid quantification of parasitemia in fixed malaria cultures by flow cytometry. Experimental Parasitology. 62 (2), 275-282 (1986).
- Jacobberger, J. W., Horan, P. K., Hare, J. D. Analysis of malaria parasite-infected blood by flow cytometry. Cytometry:The Journal of the International Society for Analytical Cytology. 4 (3), 228-237 (1983).
- Bei, A. K., et al. A flow cytometry-based assay for measuring invasion of red blood cells by Plasmodium falciparum. American Journal of Hematology. 85 (4), 234 (2010).
- Montes, M., Jaensson, E. A., Orozco, A. F., Lewis, D. E., Corry, D. B. A general method for bead-enhanced quantitation by flow cytometry. Journal of Immunological Methods. 317 (1-2), 45-55 (2006).

