Imaging CD19+ B Cells in an Experimental Autoimmune Encephalomyelitis Mouse Model using Positron Emission Tomography
Summary
This paper details methodology for radiolabeling a human-specific anti-CD19 monoclonal antibody and how to use it to quantify B cells in the central nervous system and peripheral tissues of a mouse model of multiple sclerosis using in vivo PET imaging, ex vivo gamma counting, and autoradiography approaches.
Abstract
Multiple sclerosis (MS) is the most common demyelinating central nervous system (CNS) disease affecting young adults, often resulting in neurological deficits and disability as the disease progresses. B lymphocytes play a complex and critical role in MS pathology and are the target of several therapeutics in clinical trials. Currently, there is no way to accurately select patients for specific anti-B cell therapies or to non-invasively quantify the effects of these treatments on B cell load in the CNS and peripheral organs. Positron emission tomography (PET) imaging has enormous potential to provide highly specific, quantitative information regarding the in vivo spatiotemporal distribution and burden of B cells in living subjects.
This paper reports methods to synthesize and employ a PET tracer specific for human CD19+ B cells in a well-established B cell-driven mouse model of MS, experimental autoimmune encephalomyelitis (EAE), which is induced with human recombinant myelin oligodendrocyte glycoprotein 1-125. Described here are optimized techniques to detect and quantify CD19+ B cells in the brain and spinal cord using in vivo PET imaging. Additionally, this paper reports streamlined methods for ex vivo gamma counting of disease-relevant organs, including bone marrow, spinal cord, and spleen, together with high-resolution autoradiography of CD19 tracer binding in CNS tissues.
Introduction
MS is an immune-mediated neurological disorder; the unique presentation in each patient can make management challenging for both patients and clinicians1. The disease itself is characterized by the presence of demyelinating lesions and immune cell infiltration in the brain and spinal cord, resulting in physical and cognitive impairment2. The traditional paradigm that MS is a T cell-mediated disease was first challenged in a landmark phase II clinical trial of rituximab3, a therapy targeting the CD20+ subset of B cells. Additional B cell therapies have since been developed that target CD194, a pan B cell biomarker that is expressed on a broader range of B cells, which can be both diagnostically and therapeutically advantageous. Moreover, existing methods to assess treatment efficacy (i.e., monitoring the number of relapses and magnetic resonance imaging [MRI] activity) do not afford early measures of response-thus placing patients at significant risk of CNS damage due to suboptimal therapy selection and optimization. Hence, there is a critical need for strategies to monitor specific immune cells, such as CD19+ B cells, in real time in the CNS and periphery of MS patients.
PET imaging is a robust imaging technique that allows in vivo, whole-body visualization of a given target of interest, such as CD19. While blood draws, records of relapse rates, and lesion monitoring via MRI provide snapshots into treatment efficacy, PET imaging can allow researchers and clinicians to monitor effectiveness of a therapy across the entire body. This proactive approach to therapeutic monitoring allows clinicians to assess medication effectiveness in real time, enabling rapid adjustments as needed. Monitoring the location and density of cell populations associated with disease also permits longitudinal assessment of severity using patient-specific anatomic information. It is thus essential to establish reproducible analytical methods to reliably utilize the full potential of PET imaging in clinical and preclinical settings.
This paper describes methods (Figure 1) to perform PET imaging, ex vivo gamma counting, and autoradiography (ARG) of CD19+ B cells with a 64Cu-labeled anti-human CD19 monoclonal antibody (mAb), known as 16C4-TM (64Cu-hCD19-mAb), in the experimental autoimmune encephalomyelitis (EAE) mouse model of MS induced in transgenic mice expressing human CD19 (hCD19) using human recombinant myelin oligodendrocyte glycoprotein 1-125 (MOG1-125). We also provide methods to assess radiotracer binding accurately and reproducibly in the brain and spinal cord, both critical sites of pathogenesis often severely affected in this and other neurodegenerative models. These techniques allow for non-invasive investigation of the role of B cells in disease pathology and have the potential to be clinically translated to assess the efficacy of anti-B cell therapies in MS.
Protocol
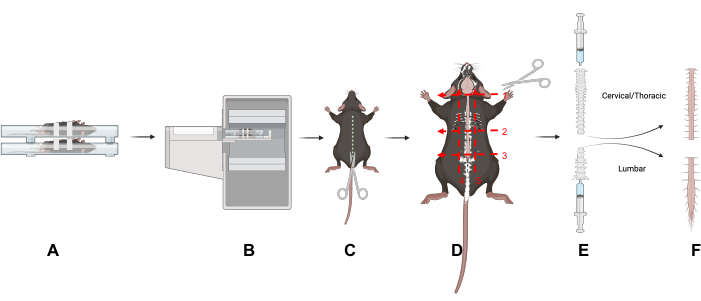
Figure 1: Study design. An overview of key techniques in this article. (A) Placing the mice in the scanning bed on their backs reduces motion in the spine. (B) PET/CT imaging of the mice. (C) Make an incision down the dorsal side of the animal to expose the spinal column. (D) Bisect the spinal column into cervical/thoracic and lumbar portions and remove the sections following the five indicated cuts. (E) Use a syringe to remove the spinal cord from the spinal column by making a seal with the syringe and spinal column and flushing from the cranial and caudal ends of the spinal column as shown. (F) Isolated cervical/thoracic and lumbar spinal cord segments. Abbreviation: PET/CT = Positron emission tomography/computed tomography. Please click here to view a larger version of this figure.
All animal studies were carried out in accordance with the Administrative Panel on Laboratory Animal Care (APLAC) at Stanford University, a program accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC International). Mice were acclimated to the vivarium for at least 7 days prior to start of study to minimize stress on the mice, as stress can affect EAE induction.
1. EAE induction in female humanized CD19 mice
- Induce humanized CD19 C57BL/6J female mice aged 9-13 weeks with EAE as previously described5 using MOG1-125.
2. Animal care and scoring in EAE mouse model
- Score disease progression and care for the mice as previously described5. In brief, score this model on a scale of 1-5 as follows: 1 is tail weakness/limpness, 2 is weakened hindlimbs, 3 is hindlimb paralysis, 4 is hindlimb paralysis with forelimb weakness, 5 is moribund.
3. mAb conjugation, radiolabeling, and characterization
- Conjugate the bifunctional chelator 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid mono-N-hydroxysuccinimide ester (DOTA-NHS-ester) to hCD19-mAb to radiolabel with 64Cu.
- Using a desalting column, exchange the hCD19-mAb storage buffer with HEPES buffer (pH 8.5-9): condition the desalting column(s) with HEPES. Calculate the number of columns needed based on desalting column sample volume capacity and volume of mAb required: 0.5 mL tubes: 30-130 µL of sample volume, 300 µL of wash; 2 mL tubes: 200-700 µL sample volume, 1 mL wash.
- Cool the centrifuge to 4 °C for buffer exchange via the desalting column.
- Retrieve the desalting column from the fridge. Remove the bottom of the desalting column, loosen the cap, and place the column in a collection tube.
- Centrifuge at 1,500 × g for 2 min to remove the storage solution; discard the flowthrough containing the storage solution and reuse the collection tube. Mark a line on the column where the highest point of the desalting compacted resin is slanted upward.
- Add HEPES buffer to the lower side of the desalting column. Place the desalting column in the centrifuge with the line facing outward; spin at 1,500 × g for 2 min. Discard the flowthrough and reuse the collection tube.
- Repeat steps 3.1.4 and 3.1.5 2x using the same collection tube and discarding the flowthrough between steps.
- Place the conditioned desalting column into a new collection tube and label; this tube will contain the hCD19-mAb.
- Add hCD19-mAb to the top of the conditioned desalting column(s) and use HEPES buffer to rinse the empty mAb vial; add this to the top of the desalting column (total volume as per the manufacturer's recommendation). Spin at 1,500 × g for 2 min to elute the hCD19-mAb. Keep the flowthrough containing the hCD19-mAb.
- Measure the concentration of hCD19-mAb using a UV/Vis spectrophotometer and adjust to 0.5 µg/µL with HEPES buffer if necessary.
- To the hCD19-mAb solution, add 1/50th the volume of the mAb solution of 0.5 M EDTA to make the final concentration of EDTA 0.01 M in the hCD19-mAb solution. Let the hCD19-mAb-EDTA solution stand at room temperature for 15 min.
- Remove the DOTA-NHS-ester from the freezer and allow it to come to room temperature (10-15 min). Calculate the volume of DMSO to add to the DOTA-NHS-ester to make a DOTA concentration that will allow the addition of the desired DOTA/mAb molar ratio (which is typically on the order of 1-2 DOTA/mAb).
NOTE: The volume of DMSO-DOTA-NHS-ester added to hCD19-mAb should not exceed 10% of the mAb volume. This should be done using a spreadsheet so it can be done quickly and repeatedly. - Based on the desired ratio of DOTA to hCD19-mAb, calculate the amount of DOTA-NHS-ester to add to hCD19-mAb.
nmol mAb × 10 DOTA/mAb → nmol DOTA → mg of DOTA → mg/mL DOTA/DMSO → mL of DMSO → dilution factor of DOTA/DMSO solution - Weigh 1-2 mg of the DOTA-NHS-ester, and carefully add the correct volume (calculated in step 3.1.11) of DMSO to the DOTA-NHS-ester; mix and spin down.
- Pipette the calculated volume of the DOTA-NHS-ester solution (step 3.1.12) into the hCD19-mAb solution; wipe the outside of pipette tip prior to addition to ensure extra DOTA-NHS-ester is not added (without changing the amount in the pipette). Gently mix and spin down.
- Place in the fridge (4 °C) to react overnight (12-16 h).
- Purification and concentration
- Cool the centrifuge to 4 °C for the centrifugal concentrator buffer exchange steps; place a metal PCR tube block on dry ice for snap-freezing the conjugate.
- Remove the DOTA-hCD19-mAb reaction from the fridge and quench by adding biological grade TRIS buffer: 10% of total reaction volume. Remove 10-20 µg of the sample for mass spectrometry analysis.
- Condition the desalting column(s) as described above (steps 3.1.1-3.1.5), using 0.1 M ammonium acetate buffer, pH 5.5.
- Buffer exchange the DOTA-hCD19-mAb solution to ammonium acetate (steps 3.1.1-3.1.8).
- Concentrate the DOTA-hCD19-mAb solution: add the solution to a 50 kDa molecular weight cut-off centrifugal concentrator following the manufacturer's recommendations on volume. Centrifuge at 4,000 × g for 10 min (or until the volume is reduced by 80%-90%); discard the flowthrough.
- Repeat nine more times (10 total): add enough ammonium acetate to bring the volume back to the maximum recommended volume.
NOTE: The total should be what was initially added, including what is left in the column, usually 400-450 µL for a 500 µL capacity centrifugal concentrator.- Rinse the reaction vial with ammonium acetate buffer to retrieve any residual DOTA-hCD19-mAb; add the wash to the centrifugal concentrator.
- Centrifuge at 4,000 × g for 10 min.
NOTE: The spin time may be reduced on subsequent spins if the volume is reduced to 80%-90% in less time.
- Remove the protein solution from the centrifugal concentrator. Note the total volume of the DOTA-hCD19-mAb.
- In centrifugal concentrator 2, add enough ammonium acetate buffer for a total volume of 100 µL and pipette to mix. Cap the centrifugal concentrator column and invert. Spin at 4,000 × g for 2 min to collect the solution. Transfer to a new tube.
- In centrifugal concentrator 500, use a pipette to collect the mAb solution from the centrifugal concentrator; add to a new tube.
- Measure the concentration using a UV-Vis Spectrophotometer. If the concentration is more than 2 mg/mL, dilute with ammonium acetate to 2 mg/mL.
- Aliquot 100 µg per PCR tube (approximately 50 µL); label the tubes with DOTA-hCD19-mAb, date, mass, and concentration. Spin down the vials.
- Snap-freeze the DOTA-hCD19-mAb on the chilled PCR block on dry ice (or freeze on dry ice). Once all the samples are frozen, place them in a -80 °C freezer.
- Measure the number of DOTA per hCD19-mAb by mass spectrometry. Keep a sample of unconjugated (pure) antibody from each conjugation to calculate the ratio. Use equation (1) given below; molecular weight is abbreviated MW.
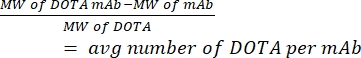 (1)
(1)
- Radiolabeling
NOTE: Wear appropriate personal protective equipment (PPE) for handling radioactivity, including a lab coat, gloves, and a personal body and ring dosimeters, following institutional regulations. Survey and change gloves regularly to prevent radioactive contamination. Use lead shielding and increase the distance from the radioactive source by handling with tongs when possible.- Transfer radioactivity from the shipping vial to a new vial using a pipette. Measure the radioactivity.
- Remove an aliquot for the first radiolabeling reaction. Add 50 µL of ammonium acetate (pH 5.5) per 1 mCi of 64Cu-CuCl3. Measure the pH by pipetting 1 µL onto a pH strip with a range that captures 5.5 with enough resolution to distinguish between 5 and 6.
- If the pH is not 4-5.5, alter it using 1 M NaOH or 0.1 M HCl. Add small amounts, 1-5 µL, of 1 M NaOH if pH is less than 4 or 0.1 M HCl if pH is greater than 5.5 until the correct pH is reached. With each addition, mix thoroughly, spin down, and measure the pH as described above. Note each addition and removal of any solution (including to check the pH) so the final volume can be calculated.
- Once the optimal pH has been achieved, add 10 µg of DOTA-hCD19-mAb per 1 mCi of 64Cu-CuCl3. Mix gently and spin down briefly.
- Place the reaction vial on a thermomixer set to 37 °C and 400 revolutions per minute (rpm).
- After 30 min, quench the reaction: divide the total reaction volume by 50, and add that volume of 0.5 M EDTA to the reaction mixture.
- Determine the labeling efficiency using instant thin layer chromatography (ITLC) to measure the percentage of bound and free copper in the solution.
- Cut 1 cm-wide strips of TLC paper. Mark 1 cm from the top and bottom of the strip and prepare a tube (50 mL conical flask) with mobile phase less than 1 cm of 0.1 M citric acid (the level of mobile phase should be below the 1 cm mark on the strip when placed in the tube).
- Pipette 1 µL of the reaction solution on the strip at the bottom 1 cm mark (solvent front) and place the strip in the tube. Watch until the solvent front reaches the top 1 cm mark, remove the strip, and wrap it in a piece of plastic wrap.
- Place the strip on the platform and run it through a radio-TLC imaging scanner. Look for the radiolabeled mAb at the origin and free 64Cu that travels with the mobile phase front (Figure 2). If the percentage of free copper is less than 5% (95% labeling efficiency), proceed with injection into animals. If the percentage of free copper is greater than 5%, proceed with purification.
NOTE: The percentage of free copper generally depends on the ratio of DOTA-mAb to 64Cu, reaction time, pH, and temperature. The reaction conditions should be optimized by each user for each new mAb to ensure consistent results.
- Purification via a disposable DNA grade gravity column
- Condition a disposable DNA grade gravity flow column (desalting/buffer exchange column) according to the manufacturer's directions. Place the disposable DNA grade gravity column on a ring stand or other instrument behind adequate lead shielding; ensure the apparatus is stable and easily movable. Place the tube underneath the column.
- Pipette the crude reaction mixture onto the gravity flow column resin. Wait until all the liquid has flowed into the resin. Pipette sufficient PBS to bring the entire volume (crude product plus PBS) to 500 µL. Dispose of the flowthrough.
- Place the column over 1.5 mL centrifuge tubes labeled 1-10. Add 1 mL of PBS to the column. Collect five drops in each tube until the flow has stopped.
NOTE: Fewer than 10 tubes may be required. - Cap the bottom of the column and measure the residual radioactivity. Measure each vial with flowthrough. For each vial with more than 50 µCi, prepare an ITLC per step 3.3.7 to measure the percent bound copper in each fraction.
NOTE: The first one or two fractions will contain only mobile phase; the radiolabeled mAb will elute first (typically fractions 2 or 3 through 5 or 6) and the free 64Cu will elute last (some will stick to the column). The percent bound 64Cu may vary between fractions. - Combine the fractions with greater than 95% binding (less than 5% free copper); use the solution for injection. If desired, perform size-exclusion HPLC to confirm radiolabeling5 and calculate the molar activity. Save an aliquot to measure the concentration on a UV-Vis spectrophotometer after it has decayed 10 half-lives (127 h or 5.3 days).
4. Dose preparation
NOTE: Before handling the dose, wear proper PPE, including lab coat, body and finger dosimeters, and gloves.
- Inject 64Cu-DOTA-hCD19-mAb 18-24 h prior to PET scanning, to allow adequate circulation of the radiotracer in the body.
- Immediately place the lead container with the 64Cu-DOTA-hCD19-mAb behind lead shielding. Turn on the Geiger counter to monitor potential contamination.
NOTE: Change gloves frequently when handling radioactive material. Double-gloving while drawing doses is recommended to facilitate swift glove changes. Always place contaminated sharps and trash in the designated shielded trash area. - Dilute the 64Cu-DOTA-hCD19-mAb to an appropriate concentration in a low bind 1.5 mL plastic centrifuge tube.
NOTE: The 64Cu-DOTA-hCD19-mAb will bind to plastic if it is not low bind; 64Cu will bind to glass.- Dilute the radiotracer in saline to prevent radiolysis and simplify drawing consistent doses.
- Prepare doses to administer between 75 and 150 µCi in 100 µL, if possible. Ensure that the maximum total injected volume does not exceed 10% of the mouse's body weight.
- Use a 500 µL (50 cc) insulin syringe to draw the dose from the low bind plastic tube. Ensure there are no bubbles in the syringe since it will be injected intravenously.
- Prelabel the syringes with the corresponding animal numbers.
- Record the dose activity and time in a lab notebook for data analysis.
- Have the doses ready for injection as soon as the animals are catheterized to reduce time under anesthesia.
- After the doses have been prepared, prepare a standard (phantom) for scanning to generate a calibration factor.
- Fill a 15 mL conical tube with 50-75 µCi of activity diluted in water (or PBS).
NOTE: Ensure thorough mixing of the solution. The standard can be free 64Cu left over from labeling.- Measure the amount of activity in the standard and record the time.
- Fill a 15 mL conical tube with 50-75 µCi of activity diluted in water (or PBS).
5. Cannulation and Injection
NOTE: See previously described methods6 for intravenous cannulation of mice for injection of the radiotracer6.
- Weigh and score the mice for disease severity as described in section 2.1 before anesthetizing mice in a knockdown box filled with 1.5-3% isoflurane in preparation for radiotracer administration.
NOTE: These mice will be injected on the bench top, not on the PET scanner as previously described. There is no need to glue the catheters in place for injection, as the mice will not be moved between cannulation and injection. - Once a mouse is cannulated, insert the needle into the end of the catheter and inject slowly. After injection, follow with a small saline flush through the catheter to ensure the entire dose is injected.
NOTE: The volume should approximately equal the dead volume of the catheter, which is 50 µL for the catheters used by the authors.- Inject over a piece of laboratory wipe to collect any drips of radiotracer; include this when measuring the residual dose.
- Record the time of injection in a lab notebook.
- Remove the cannula immediately after injection. Measure the cannula, dose syringe, and tissue using a dose calibrator to determine the residual dose that was not injected into the mouse.
- Record the activity and time in a lab notebook.
- After the mice are injected, label their cages with a cage card indicating the mice are radioactive. Record and log the cages according to institutional guidelines. Then, place the mice in a designated radioactive holding area.
6. PET/CT imaging
- Perform PET imaging 18-24 h after the injection of 64Cu-DOTA-hCD19-mAb. Weigh and score the mice prior to scanning.
NOTE: The scanner operating instructions depend on the scanner being used.- Ensure the X-ray component of the PET/CT scanner is warmed up and ready for acquisition.
- Open the PET scanner acquisition software on the computer.
- From the Investigator Login dropdown menu, click on the appropriate lab information.
- On the Project page, either create a new project or select an existing project from the dropdown menu.
- When the Initialization prompt automatically appears on the screen, click on Home Bed and wait for bed to home. Then, click on Warm up CT.
- Ensure the CT shielding door is closed to allow the CT to warm up.
- While the scanner is warming up, score the EAE mice and inject them subcutaneously with 0.2-0.4 mL warm saline in the flank for hydration.
- After the scanner is warmed up, go back to the computer to set up the PET scans for the study.
NOTE: These can be set up prior to the day of scan.- Under the Recent Studies header, click on Create New Study. Fill out the study name, protocol, compound, and subject information.
- If performing a PET/CT, select PET Protocol first, and then select CT Protocol.
NOTE: Typically, a standard CT is sufficient for scanning the EAE mouse model. A CT scan is 1 min long and acquired with binning at 2 x 2 at voltage 80 kVp, current 150 µA, and 720 projections. CT images are reconstructed using a Modified Feldkamp Algorithm. - For the PET protocol, choose a 10-15 min static 64-copper scan. If this scan is not already on the list of available protocols, add it by clicking on the Protocols tab in the Standard menu, Create New Protocol | Isotopes dropdown menu. If the desired isotope is not listed, click on More | Add from Library and add the desired isotope. Define the scan duration, click on the radio button for Static Scan, name the protocol and click on Save.
- Return to the Studies tab and continue to name and set up all the desired scans for the day.
NOTE: It is recommended to also set up one "CT Test" scan using only a standard CT to check the bed positioning of the first scan to ensure optimal placement. This should be the first scan run for the study.
- If performing a PET/CT, select PET Protocol first, and then select CT Protocol.
- Under the Recent Studies header, click on Create New Study. Fill out the study name, protocol, compound, and subject information.
- Once the scanner is ready, anesthetize the mice in a knockdown box to prepare them for the scan.
NOTE: At this stage, the mice are likely very sick; it is best practice to minimize the time under anesthesia.- Apply eye gel.
- Ensure the four-mouse scanning bed is equipped with heating elements, such as heating pads or heated air, with isoflurane set to 1.5%-2% and heating pad turned on (Figure 3). Place the mice in a supine position on the scanning bed.
- As EAE disease progresses, the spine of the mouse becomes severely curved. Scan the mice while they are on their backs to straighten the spine as much as possible, improving the analysis downstream. Gently pull the mouse tail to aid in straightening the spine.
- Once in the supine position, securely tape each mouse in place with soft microscope tape. Use one strip of tape over the head and another gently over the belly to minimize motion due to breathing.
- Record in a lab notebook which mouse is in what scanning position.
- Once the mice are secured, return to the scanning computer to operate the scanner.
- Open the Motion Controller menu. Click on PET Center FOV to move the mice on the scanning bed into the PET ring. For the first scan of the day, click on CT Center FOV. Once in position, run the CT Test scan to ensure the position is correct; repeat until the bed position is satisfactory.
- Place a small piece of white tape on the scanning bed to mark the correct bed placement for the remainder of the study.
- Once the bed is in place for the PET scan, initiate the scanning sequence by clicking on Run.
- Wait for the scanner to automatically move from the PET ring to the CT.
- Always visually check whether the animals have moved into the proper position for both PET and CT.
- Record the scan start time in a lab notebook for decay correction of the dose injected during data analysis.
- After the scan is complete, allow the image to reconstruct. Check the data before removing the animals.
NOTE: 3D-ordered subsets expectation-maximization (OSEM) reconstruction will take approximately 5 min for a static scan. - Visually check and approve the data, using the spleen as a positive control as this tissue contains a large number of B cells. Remove the animals from the scanning bed and place them in an isoflurane-filled knockdown box in preparation for perfusion and subsequent dissection.
- Repeat steps 6.5-6.12 for the remaining mice in the study.
- When all the mice are scanned or when scanning a group that has an open place in the scanning bed, scan the standard prepared in step 4.6.
7. Dissection for ex vivo gamma counting and autoradiography
- Before dissecting, ensure all gamma counting and centrifuge tubes have been pre-weighed.
- Perform euthanasia via perfusion, as previously described6, with PBS and thoracotomy while the mice are deeply anesthetized (continual inhalation of 4% isoflurane, 2 L/min 100% O2).
- To remove the bone marrow, cut both femurs at the knee and pelvis. Make sure both heads are removed from the femur.
- Place both femurs in a 0.5 mL centrifuge tube that has a hole at the bottom (using a 20 G needle) and has the lid cut off.
- Place the 0.5 mL tube containing the femurs inside a 1.5 mL centrifuge tube with the lid cut off.
- Place the entire tube setup in a mini centrifuge. Spin for 4 min at 4,500 × g.
NOTE: The bone marrow should be dislodged through the hole in the 0.5 mL centrifuge tube and settle at the bottom of the 1.5 mL centrifuge tube.- Separate the tubes. Weigh the empty femurs in the 0.5 mL centrifuge tube. Weigh the bone marrow in the 1.5 mL centrifuge tube. Place each centrifuge tube in gamma counting tubes.
- Remove the brain using forceps and scissors, taking care to keep the brainstem intact. Place the brain in a gamma counting tube. Record the dry weight, flush with PBS, and keep on ice until ready for counting.
- To remove the spinal cord, perform the following steps.
- Remove the skin and fur by making an incision down the dorsal side of the animal to expose the spinal column (Figure 1).
- Separate the lumbar (L) from the cervical (C) and thoracic regions by cutting along three transverse planes around and through the spinal column: at the base of the neck (C1 vertebra) (Figure 1D, Number 1); at the base of the ribcage (L1 vertebra) (Figure 1D, Number 2); at the base of the pelvis (L5 vertebra) (Figure 1D, Number 3).
- Cut under the rib cage (Figure 1D, Number 2).
- Cut directly above the sacrum to separate the lumbar spinal region. Carefully trim the spinal column from the pelvic end until the lumbar spinal cord is visible (Figure 1D, Number 3). Trim off the surrounding tissues to isolate the lumbar and cervical/thoracic regions of the spinal column (Figure 1D, Numbers 4 and 5).
- To expel the spinal cord, use a slip-tip syringe (3-10 mL) filled with PBS. Create a seal between the syringe and the spinal column using the thumb and forefinger.
- Gently push the PBS through the syringe to expel the spinal cord onto an absorbent pad (Figure 1E); repeat for both spinal regions. Place the spinal cord tissues in a gamma counting tube.
- Record the dry weight, and add PBS to ensure the tissue is at the bottom of tube to avoid drying. Place the tube on ice until ready for counting.
- Expel the cervical/thoracic spinal cord from the cranial end and the lumbar spinal cord from the caudal end of the spinal column (Figure 1E).
8. Ex vivo gamma counting
- Open the gamma counter software. Navigate to the work list and select the desired protocol, such as a 30 s counting protocol for 64Cu.
- Prepare at least three standards in separate tubes. Run them now for use in analysis (step 10.2). Aim to make three replicate volumes and amounts of activity in three separate tubes.
NOTE: A volume of 500 µL gives good results. While activity will be determined by the machine used, 10 µCi generally works well. - Place the standards in a rack labeled with the barcode corresponding to the desired protocol to be run. Place the rack on gamma counter.
- After recording the organ weights, place the tubes containing the organs on the gamma counting rack after the tubes containing the standards.
NOTE: Organs of interest for this model can include axial lymph nodes, blood, bone marrow, brain, cervical lymph nodes, femur, heart, liver, lumbar spinal cord, muscle, spleen, tail, and cervical/thoracic spinal cord. - Put a rack with a stop barcode in the back of the gamma counter.
- Press the Play button on the computer. If possible, do not press Play until there are multiple racks or organs to be run to allow all tubes to be continuously counted in one file. Ensure that a rack with a stop barcode is in the back of the gamma counter for each run.
- Run until all the samples have been counted. Save and export the file.
9. Ex vivo autoradiography (ARG) of CNS tissue
- Follow previously published steps for both brain and spinal cord ARG (while excluding steps 2-6 described by Chaney et al., as the mice are already injected with radiotracer from the PET scan)6.
NOTE: Specific instructions for preparing the spinal cord ARG cassette are listed here6. - After gamma counting is complete for the lumbar and cervical/thoracic spinal cords, promptly place the tubes on ice until all CNS tissues have been counted.
NOTE: Refer to the previously published method for ARG of the brain6. - Gently tip the tubes to allow the spinal cords to fall out onto an absorbent pad. If the spinal cord sticks to the side of the tube, gently flush with cold PBS and tip the tube again. Carefully dry each spinal cord by gently blotting with a lab tissue. Place the dried spinal cords in an organized fashion on a thick black piece of paper.
- Label next to the spinal cords with a white pen for easy identification.
- Leave space at the corners and in the middle of the black paper to place stacks of three microscope slides to act as spacers to prevent the compression of the spinal cords when the ARG cassette is closed. Use 5-7 stacks.
- Once all the lumbar and cervical/thoracic spinal cords are positioned on the black paper and labeled, carefully place the piece of paper in the ARG cassette. Place the open cassette on a tray of dry ice to freeze the spinal cords.
- Once frozen, gently place plastic wrap between the digital phosphor storage screen and the spinal cords and place the screen on top of the samples. Immediately close the cassette and place it in the -20 °C freezer for approximately 10 half-lives (~127 h).
- When the exposure time is complete, scan the film using a phosphor imager. Analyze the resulting digital image (see section 12 for instructions).
10. Analysis of biodistribution data
- Set up a "Dose Correction" spreadsheet to mathematically determine the time correction for the radioactive decay, thus normalizing radiation doses and allowing comparisons between subjects.
- Decay-correct all doses to the injection time, accounting for residual activity left in the syringe and catheter after injection.
- Using the standards prepared in step 8.2, average the activity amount (µCi) and normalized counts per minute (CPM). Divide the average CPM by the average standard activity amount to get CPM/µCi.
NOTE: Ensure the activity amount for each standard is decay-corrected to the time the gamma counter counts the CPM of the standards. The gamma counter should normalize all CPM values to the start time of the protocol.
- Set up the "Results" spreadsheet to calculate the final percent injected dose per gram of tissue (%ID/g) for each sample.
- Decay-correct the normalized CPM from the gamma counter for each sample counted to the injection time of the animal.
NOTE: Decay correction can be to any time point; ensure all doses and CPM values are decay-corrected to the same time point. - Normalize the decay-corrected CPM to the mass of each sample to determine the CPM per sample. Calculate the total CPM injected by subtracting the CPM in the tail from the calculated injected CPM.
NOTE: The tail does not need to be mass-corrected as this CPM value will simply be subtracted from the total injected CPM to account for any extravenous tracer from injection. - Divide the CPM per mass by the total CPM injected to calculate %ID/g.
- Decay-correct the normalized CPM from the gamma counter for each sample counted to the injection time of the animal.
- Set up a "Summary" spreadsheet to display the final results for input into a graphing software and subsequent data visualization and statistical analysis.
11. PET image analysis
- Open the PET analysis software. lick on File |Open Local Data | DICOM. Locate the desired file (DICOM format). Open both the PET and CT.
- In the Data Manager, adjust the PET and CT contrast to the desired levels.
- Register and crop individual mice.
- In the Navigation menu, select the Reorientation/Registration tab.
- Go to the Rigid menu within this tab. Designate the CT scan (0) as the Fixed scan and the PET scan (1) as the Moving scan.
- Select Rigid Transformation and Fast Quality. Click on Registrar.
- After registration is complete (5-10 min), click on the check mark to save the registration.
- Visually inspect the data to ensure registration was successful. Export the session by clicking on File | Session | Export.
- Next, crop each mouse in a full body crop: go to the Navigation menu and click on Cropping. Drag the sides of each cross section from the outer edge inward.
- Once the desired mouse is tightly cropped, click on the check mark, and export the session to save.
- Next, crop and straighten the heads of each mouse for brain analysis using the manual Translation, Rotations, and Flips in the Registration/Reorientation menu. Export to save.
- Analyze the spinal cord.
- To begin analysis of regions of interest (ROIs) in the spinal cord, open the 3D ROI Tool from the Navigation Menu.
- Under the ROIs header, use the plus sign at the bottom of the menu to create six ROIs: Lumbar ROI, Cervical/Thoracic ROI, Lumbar Skeleton, Thoracic Skeleton, Lumbar Spinal Cord, Thoracic Spinal Cord.
NOTE: The Lumbar and Cervical/Thoracic ROIs are generalized large ROIs that will be used to create the skeleton ROIs (Figure 4). - To avoid visual interference from the PET signal with this step, click on F3 to turn off the PET.
- Go to the top of the 3D ROI Tool Operator. Click on the solid dot to the right of the cursor symbol to open the 3D Paint Mode and Erode/Dilate menu.
- Select Sphere and change the size para 20 pixels. Set Dilate para +5.
- Before proceeding, go to the bottom of the menu. Ensure the Lumbar ROI is selected, as this is the first ROI to be drawn.
- On the CT, find the L6 vertebra of the spinal column (where the spine meets the hips). Starting one vertebra above L6, draw a rough ROI Lumbar ROI over the five vertebrae above the hips (L1-L5 vertebrae). Then, switch to Cervical/Thoracic ROI and trace the remainder of the spine to the base of the skull.
NOTE: This does not have to be precise, as it is used to indicate where Otsu thresholding should occur in step 11.4.8. - After drawing the generalized ROIs, go to the top of the operator. Select the Segmentation Algorithms menu.
- From the dropdown menu, select Otsu Thresholding. For the input, select Lumbar ROI. At the bottom of the menu, ensure Lumbar Skeleton is selected. In the dropdown menu next to Image, ensure the CT scan is selected and click on Apply. Repeat for Cervical/Thoracic ROI and Thoracic Skeleton.
- If the Otsu Thresholding does not sufficiently highlight the spinal column, use Global Thresholding and change the Min value to 350 and the Max para 3,500 for manual thresholding and adjust as necessary to isolate the vertebrae.
- After using Otsu Thresholding to create the Skeleton ROIs, return to the Navigation menu (cursor icon). Either delete or checkmark the H (hide) column for both the rough Lumbar and Cervical/Thoracic ROIs to hide them. Checkmark the I (immutable) column for both Skeleton ROIs so they cannot be edited.
- Finally, return to the top of the 3D ROI Tool Operator and go to the 3D Paint menu to draw the Spinal Cord ROIs.
- Select the Sphere tool again and trace the spinal cord within the skeleton for both Lumbar and Thoracic, ensuring the correct ROI is selected at the bottom of the menu.
- To erase any ROI, click on Command/Control and draw over the part to be erased.
- Check the spinal cord ROI from all three planes to make sure there is no ROI drawn outside of the spinal column.
- Export spinal cord analysis results.
- If the PET signal was turned off in step 11.4.3, press F3 after the Spinal Cord ROIs are drawn to turn the PET back on, or select the Visual Controller (VC) and click on the PET bar.
- Go back to the Navigation menu (cursor icon). Click on the grid icon to Show Table. Copy the table into spreadsheet software and save.
- Lastly, export the file in PET analysis software, as described above, to save the ROIs drawn.
- Analyze the brain using a semi-automated 3D atlas.
- Open the head crop file. Import the mouse brain atlas by going to the Advanced Modules menu and selecting the 3D Brain Atlas Tool. Make sure the reference is set to CT and the crop option is unchecked. Set a pathway for the output directory.
- In Advanced Setting, change Transform para Versor-Affine. Keep all other default settings. Click on Run.
- Manually adjust the atlas in the Reorientation/Registration menu and use the CT of the skull as a guideline for fitting the atlas.
- Use great care if scaling is necessary, as this can greatly affect brain structure volumes. Click on the checkmark when adjustment is complete.
- Rerun the atlas with Import 3D ROIs selected.
- Export the file to save the atlas fitted to the cropped head.
- After drawing and exporting all ROIs from the desired organs, calculate a standard binding correction value. Decay-correct all data and convert to %ID/g as previously described6. Normalize to the organ that suits the animal model, such as the heart to normalize to radiotracer present in the blood pool.
12. Ex vivo autoradiography analysis
- Open the digital image file (.gel) using the image analysis software. Adjust brightness and contrast to the desired threshold. Apply a suitable color lookup table if desired.
NOTE: Royal or Grays are recommended for ease of visualization.
Representative Results
The hCD19-mAb was DOTA-conjugated and radiolabeled with 64Cu as shown in Figure 2. EAE and naïve mice underwent PET/CT scanning (Figure 3) 18-24 h after injection with 64Cu-DOTA-hCD19-mAb. PET/CT images were co-registered using the PET analysis software, and the CNS tissues were analyzed using manual ROIs or a semi-automated 3D brain atlas. Radiotracer binding in ROIs (Figure 4) was higher in EAE mice than in naïve mice. Ex vivo gamma counting and ARG showed increased binding in the spinal cord (both lumbar and cervical thoracic segments) and brain (ARG only) of EAE mice compared to naïve (Figure 5 and Figure 6). Ex vivo gamma counting of perfused mice also showed decreased radiotracer binding in peripheral organs, including spleen, femur, and bone marrow (Figure 5), consistent with B cells leaving the periphery and infiltrating the CNS in this EAE model.
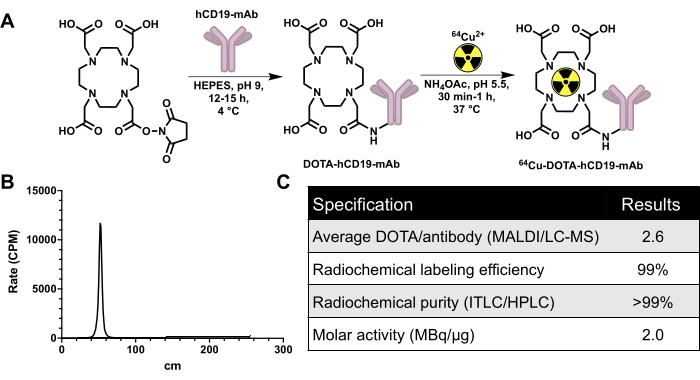
Figure 2: Conjugation and radiolabeling scheme for generating 64Cu-labeled human-specific CD19 monoclonal antibody, 16C4-TM mAb (64Cu-DOTA-hCD19-mAb), in addition to quality control data. (A) Reaction of DOTA-NHS-ester with hCD19 monoclonal antibody to produce hCD19-DOTA conjugate (not to scale) and radiolabeling with 64Cu-CuCl3 to produce 64Cu-DOTA-hCD19-mAb. (B) Representative ITLC chromatograph. The peak at 40-60 cm is the radiolabeled antibody; unbound 64Cu-CuCl3 travels with the mobile phase and would be present from 200 to 240 cm. There is no detectable free 64Cu-CuCl3 in this chromatograph. (C) The quality control specifications of the radiolabeled antibody. Abbreviations: DOTA-NHS ester = 1,4,7,10-Tetraazacyclododecane-1,4,7,10-tetraacetic acid mono-N-hydroxysuccinimide ester; ITLC/HPLC = instant thin layer chromatography/high-performance liquid chromatography; MALDI/LC-MS = matrix-assisted laser desorption/ionization/liquid chromatography-mass spectrometry; CPM = counts per minute. Please click here to view a larger version of this figure.
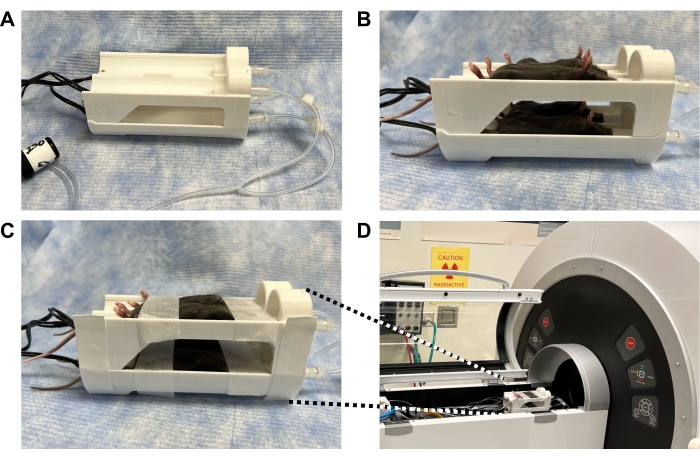
Figure 3: Photographs demonstrating how to secure mice in a 3D-printed bed within the PET scanner to enable high quality imaging of the spinal cord and brain while minimizing motion. (A) 3D printed four-mouse scanner bed (also known as "mouse hotel") equipped with heating elements and anesthesia tubing. (B) Anesthetized mice in a supine position to maximize straightness of the spine; the bed position of each mouse is recorded. (C) Mice taped securely across their head to minimize motion in the brain and across the belly to minimize motion from breathing, without affecting breathing. (D) Mouse bed positioned within the scanner and taped to the scanning bed. Anesthesia tubing was connected from scanner to bed and isoflurane set to 2%. Mouse breathing was monitored to ensure appropriate isoflurane level before closing the scanner door. Abbreviation: PET = Positron emission tomography. Please click here to view a larger version of this figure.
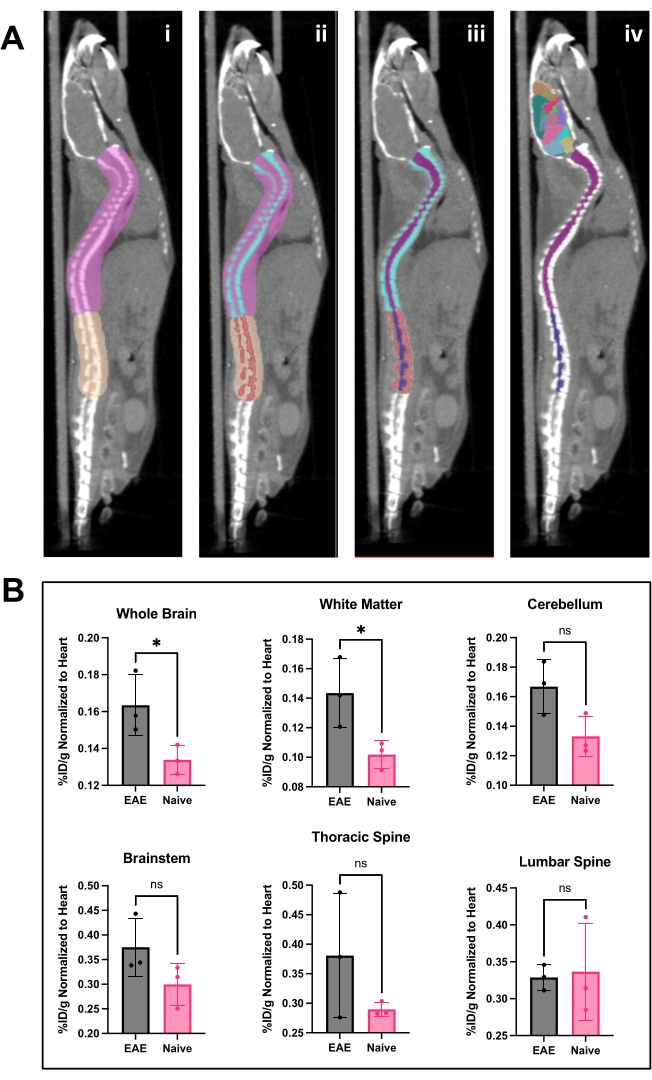
Figure 4: Spinal cord image and brain analysis and results using PET analysis software. (A) i) ROIs (pink and tan) drawn on spine to separate lumbar from the thoracic and cervical vertebrae and prepare image for Otsu thresholding. ii) Spinal vertebrae (turquoise and red) were segmented out using Otsu Thresholding. iii) Vertebrae were then made immutable in the 3D ROI menu, and the spinal cord divided into cervical/thoracic (purple) and lumbar (navy) ROIs. iv) The vertebral ROI was removed, leaving spinal cord ROIs and representative brain atlas applied. (B) Representative analysis of PET results from various CNS regions represented as %ID/g normalized to the ROI of the heart within each animal. PET acquisition was a 10 min static scan via PET/CT imaging. Brain regions quantified using a semi-automated brain atlas approach, shown in panel A. iv) Representative results show either significance or trending toward significant increase in tracer binding in the brain and thoracic spinal cord. Statistics performed using Student's t-test (*: p < 0.0332). Abbreviations: PET = Positron emission tomography; ROIs = regions of interest; CNS = central nervous system; CT = computed tomography; %ID/g = percent injected dose per gram of tissue; EAE = experimental autoimmune encephalomyelitis. Please click here to view a larger version of this figure.
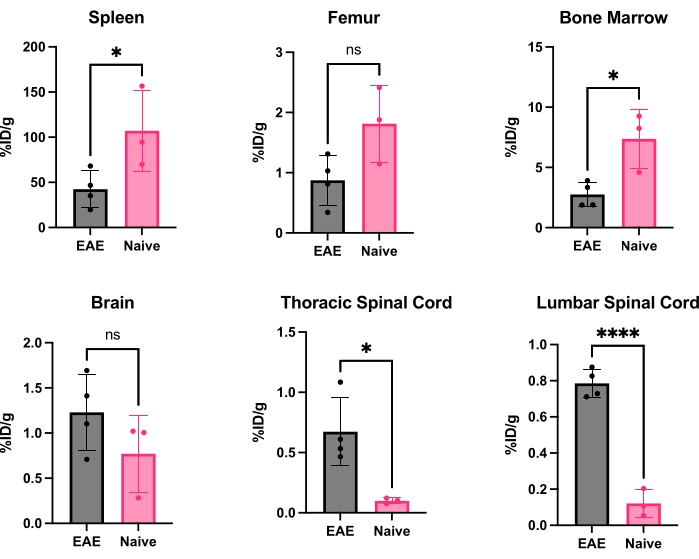
Figure 5: Representative quantification of ex vivo gamma counting in various organs in EAE and naïve mice expressed as %ID/g. Post-PET scan, mice were perfused with PBS to remove the radiotracer present in the blood, either free or bound to blood-resident CD19+ B cells, and organs quickly dissected and weighed to have an accurate weight of each organ. Tracer binding is significantly decreased in the spleen and bone marrow in EAE mice compared to naïve ones. Increased radiotracer binding is observed in both the lumbar and cervical/thoracic spinal cord segments of EAE mice. The brain does not show significant increase in radiotracer signal, though it is trending toward significant increase. Statistics performed using Student's t-test (*: p < 0.0332; ****: p < 0.0001). Abbreviations: PET = Positron emission tomography; %ID/g = percent injected dose per gram of tissue; EAE = experimental autoimmune encephalomyelitis; PBS = phosphate-buffered saline. Please click here to view a larger version of this figure.
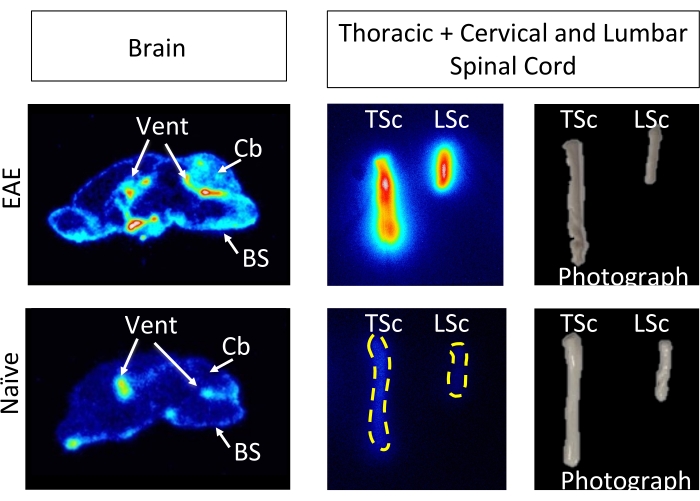
Figure 6: Ex vivo ARG images depict 64Cu-DOTA-hCD19-mAb binding in sagittal brain sections and whole spinal cords from EAE compared with naive mice. Digital phosphor storage films were scanned using a phosphor imager after being exposed to radioactive tissue samples for approximately 10 half-lives (127 h or 5 days). The resulting images reveal visually higher signal in the brain of EAE mice compared to brain sections from naïve mice, which is expected due to the regions known to contain B cells in this model5. Specifically, there is increased tracer signal in the brain stem, cerebellum, and ventricles of EAE mouse brain sections. This increase in signal for EAE mouse brain sections mirrors what was found for in the whole-brain PET quantification detailed above. Similarly, there is an increase in radiotracer binding in both the cervical/thoracic and lumbar spinal cord segments compared to naïve spinal cords, reflecting what was found using ex vivo gamma counting. Abbreviations: PET = Positron emission tomography; EAE = experimental autoimmune encephalomyelitis; Vent = ventricles; Cb = cerebellum; BS = brain stem; TSc = thoracic and cervical spinal cords combined; LSc = lumbar spinal cord. Please click here to view a larger version of this figure.
Supplemental Figure S1: Staining of CNS tissues of naïve and EAE mouse brain tissue with CD45R/B220. B cells are observed in brainstem, meninges, and white matter of EAE mice (n = 7 EAE, n = 5 naïve mice, average four slices per animal). This figure is from 5. Scale bars = 5 mm (low magnification [1x]) in sagittal brain images, 100 µm (high magnification [20x]) in brainstem, meninges, and cerebellar white matter. Please click here to download this File.
Discussion
This article describes a streamlined method for imaging human-CD19+ B cells in a mouse model of MS using CD19-PET. Due to the heterogeneous presentation of MS and varying responses to treatments, its management in the clinic can be challenging and new approaches for therapy selection and monitoring are greatly needed. PET imaging could serve as a powerful tool for monitoring disease progression and individual response to B cell-depleting therapy. In addition to MS, CD19-PET imaging could be used to monitor B cell depletion after treatment in subtypes of lymphomas and leukemia or other B cell mediated diseases. This protocol and representative data show the utility of imaging B cells in neurological diseases.
To study human CD19+ B cells in the context of MS, we chose the B cell-dependent MOG1-125 EAE model7. Similar to other EAE models, this model presents with symptoms of progressive paralysis and infiltration of immune cells into the CNS. However, the MOG1-125 model is unique in that it is a B cell-driven model: mice contain varying numbers of B cells in the subarachnoid space in the meninges, brain stem, parenchyma, and ventricles. These lymphocytes can be sparsely scattered throughout these regions and/or form follicle-like structures, which are also observed in humans with MS8,9. In addition to using naïve mice as controls, a complete Freund's adjuvant (CFA)-only induction kit may be used (i.e., an identical induction emulsion to what is given to EAE mice sans the MOG protein). In the EAE mouse model, the blood brain barrier (BBB) is dysfunctional and allows larger entities, such as antibodies, to cross. The CD19-mAb radiotracer will only bind and remain in the CNS if B cells are present; the tracer will circulate back into the blood pool if B cells are not present. We have demonstrated this using gamma counting and ex vivo autoradiography of CNS tissues by perfusing before measuring the radioactivity levels in the tissues. We have also demonstrated this in earlier publications reporting the use of mAb-based PET radiotracers (i.e., immunoPET imaging approaches) for detecting B cells in the CNS1,2.
The DOTA chelator was used since it has been used in clinical PET imaging with copper-64 labeled peptides and antibodies, and we aim to translate the hCD19-mAb for clinical imaging of MS patients. DOTA has adequate binding affinity to copper-64 in vivo. The in vivo stability is very important because free 64Cu goes to the liver and can obscure the signal of bound radiotracer; thus, it is important to measure the signal in the liver to calculate the relative signal compared to other organs. Muscle is typically taken as a control tissue, but in the case of EAE, there can be inflammation present in the muscles. The half-life of 64Cu is 12.7 h, which affords ample time for the DOTA-hCD19-mAb to bind to its target while ensuring signal can be measured by PET. When preparing the conjugate, small-scale (75-125 µg) test reactions should be performed to determine the amount of DOTA to add to mAb to produce the desired DOTA/mAb ratio (e.g., a reaction of 6-10-fold excess DOTA-NHS-ester per mol mAb may afford a conjugate of 1-2 DOTA/mAb). The reaction time and temperature (e.g., 2-4 h or overnight at 4 °C or room temperature) also influences the DOTA/mAb ratio and should be optimized. A titration with nonradioactive copper can be performed to calculate the number of DOTAs per mAb; however, we recommend performing MALDI-MS and/or LC-MS for more reliable and accurate results.
The calculated DOTA/mAb ratio is an average value for a particular sample and some variation is expected. For MALDI, several shots are taken per sample for the conjugated and unconjugated mAbs. We then calculate the ratio of conjugated to unconjugated to determine the average number of DOTA/mAb. The DOTA/mAb ratio is important because too many chelators will disrupt antibody binding and too few will lead to inconsistent radiolabeling and low signal. The ratio should be very close between batches of conjugate to maintain consistent signal intensity and binding kinetics; ideally, the same batch of conjugate should be used for all experiments within a particular study. A promising technique to reduce potential effects on immunoreactivity due to possible overconjugation is to use site-specific conjugation10 whereby the chelator conjugation is site-selective on the heavy chain glycans of the antibody, thus guaranteeing the addition of 1 chelator per mAb.
The radiolabeling reaction conditions should be optimized to ensure the highest labeling efficiency and yield since differences in antibody, DOTA/mAb ratio, and 64Cu molar activity, among other conditions, will impact radiolabeling. Using the optimal 64Cu to mAb conjugate ratio may allow the radiotracer to be used without purification, reducing the time required for radiolabeling and loss due to the gravity flow column and radioactive decay. A consistent and reliable molar activity may also be achieved when the same 64Cu to mAb conjugate ratio is used, which is especially important when comparing results across multiple cohorts of mice or imaging studies. The ITLC conditions may also be modified to suit each user. If purification is necessary, an aliquot should be saved for either HPLC and/or UV/Vis spectrophotometry so that the molar activity can be calculated.
It is important to note that using radiolabeled antibodies for imaging can be challenging. It is essential that the antibody used for the radiotracer be biologically inert so as not to have a physiological effect. Moreover, since antibodies have a long blood residence, one must wait long enough for circulation, binding, and clearance of a given mAb to ensure a suitable signal-to-background without compromising image quality. Typically waiting for 20-48 h for a 64Cu-labeled mAb is sufficient but one should image at 2, 4, 6, 12, 24, 48 h post injection when assessing a new mAb PET tracer to determine the best time point for imaging in a given rodent model. The same is true for acquiring ARG images with the highest signal-to-background ratio. The representative images in this protocol were taken at 18-20 h post injection, though other time points may be used depending on the radioisotope used. Different antibodies binding to different epitopes of CD19 will produce varying results and should be rigorously characterized.
When analyzing the spinal cord signal, it is important to position mice on their backs in the scanning bed to reduce movement caused by breathing. Additionally, supine placement can help straighten the spine in mice that have increased spinal curvature due to the progression of EAE disease. Another important aspect to consider when aiming to detect signal in the spine and spinal cord is to avoid injecting MOG1-125 on the flank as the injection sites can bind the tracer due to the associated immune response in those areas. The close proximity of the injection site can interfere with spinal cord analysis; thus, injections in the chest are preferable for the application described herein.
The image analysis techniques used are specific for CNS imaging. A brain atlas tool within the image analysis software affords reproducible and reliable results as long as the registration of PET and CT is accurate. Using the semi-automated 3D brain atlas and adjusting it to fit the skull of each mouse allows for consistent ROIs between animals. Since there is currently no automated or semi-automated approach for analyzing the signal in spinal cords, manual ROIs must be drawn. Notably, when quantifying CD19+ B cells (or any cell type present in both the bone marrow and spinal cord), it is critical to eliminate the signal arising from the spinal column and bone marrow as much as possible. The reason for this is that naïve mice are known to contain more CD19+ B cells in their bone marrow than EAE mice, in which B cells leave the periphery to infiltrate the CNS5,11. This bone marrow signal can obscure the true signal in the spinal cord.
To delineate true spinal cord signal while minimizing the contribution of signal from the spinal column and bone marrow, Otsu thresholding of the CT image can be used to make an immutable ROI for the spinal column. A separate spinal cord ROI can then be easily drawn within the spinal column. The same technique can also be applied to measure bone marrow in the femur. This is a very useful method to gain insights into tracer binding in the spinal cord. However, due to the relatively low spatial resolution of PET and issues pertaining to the partial volume effect when scanning small anatomical regions of mice, use of additional ex vivo confirmatory techniques (e.g., gamma counting, ARG) enables the validation of radiotracer binding in the spinal cord without the presence of blood, cerebrospinal fluid, or spillover signal from the spinal column.
Signal in the cervical/thoracic spinal cord tends to vary in the EAE mice depending on the severity of disease and how many B cells infiltrate during the adaptive immune response. This variation in the number of B cells that infiltrate, as well as the small amount of B cells in the CNS compared to those in the pelvic/spinal bone marrow of naïve mice, can make in vivo quantitation of spinal cord tissue challenging in mice. Given the spatial resolution of PET in small animal imaging, signal from the bone marrow can spill over onto the spinal cord signal. Ex vivo biodistribution and autoradiography completed here aid in validating the PET signal of the vertebrae versus spinal cord tissue. Mice are perfused prior to dissection to remove any unbound tracer in the blood pool so that the gamma counting and autoradiography results reflect the tracer that is actually bound in each organ rather than the tracer that is in the blood pool in that organ.
Radiotracers circulate through the blood, and with antibody tracers, specifically, there is often unbound radiotracer present in the blood for weeks after the initial injection. Since we are imaging the brain and spinal cord, which have many blood vessels, it is important to understand what portion of the signal is truly due to tracer binding in brain/tissue of interest versus that present in the blood pool. It is thus necessary to divide the brain signal by signal in the heart/blood pool. In the clinical setting, the same image analysis techniques of Otsu thresholding of vertebrae and ROIs of spinal cord tissues can be used for quantification. Given the larger tissue volumes in humans compared to mice, there should be significantly less impact from partial volume effects, leading to improved accuracy and negating the need for ex vivo techniques to confirm in vivo findings. The use of PET in the clinic will allow clinicians to personalize therapy for each patient depending on their individual B cell burden.
ARG is particularly useful for acquiring high-resolution images to enable more accurate delineation of the spatial location of tracer binding in small regions such as brain stem and cerebellum. The same sections and/or adjacent sections can be saved for immunohistochemical stains to confirm the presence of B cells. We have previously stained CNS tissues with CD45R/B220 (Supplemental Figure S1) to correlate the number of B cells with PET and ARG signal5,9. The staining can then be compared spatially to the ARG results to verify that the radiotracer signal matches the staining pattern. B cells can be present in clusters or diffusely throughout the brain stem; PET sensitivity is sufficiently high to measure the signal, which is encouraging for clinical translation. For spinal cord ARG, removing the spinal cord from the vertebrae ensures that the signal measured is due to tracer binding in the spinal cord tissue rather than the bone marrow and/or blood, which can obscure PET images due to partial volume effects.
Similar to ARG, ex vivo gamma counting enables the quantification of radioactive signal in individual organs. For this particular technique, it is important to measure the wet weight of tissues and ensure they are at the bottom of their respective tubes before placing the tubes in the gamma counter. The tubes must be labeled with the mouse number and tissue, so that the correct tube is used; the tube is then weighed on a calibrated balance and organs are inserted to the nearest tenth of a microgram (0.0001 mg). Some tissues are extremely small and the difference in the tube mass before and after will be in the order of 0.0001 mg. The tissues should be weighed immediately following dissection to prevent loss of moisture, which results in a lower mass. After weighing, the brain and spinal cord tubes should be filled with PBS to prevent from drying before freezing these tissues for ARG.
Declarações
The authors have nothing to disclose.
Acknowledgements
We are grateful for the support from the SCi3 small-animal imaging facility at Stanford and Dr. Frezghi Habte for his technical assistance with the PET/CT. LC-MS is performed by the core staff at the Stanford University Mass Spectrometry (SUMS) core facility and we appreciate the staff for providing this service. We thank Horizon Therapeutics for very kindly providing the hCD19-mAb and Jodi Karnell in particular for her technical guidance and support. This work was funded by the NIH NINDS (1 R01 NS114220-01A1).
Materials
| 0.5 mL 50 kDa MWCO Centrifugal filter | MiliporeSigma | UFC505008 | centrifugal filter |
| 64Cu-CuCl3 | Washington University in St. Louis; University of Wisonsin, Madison; or another vendor | ||
| AR-2000 Radio-TLC Imaging Scanner | Eckert & Ziegler | AR-2000 | |
| Autoradiography cassette | Cole Palmer | EW-21700-34 | Aluminum, 8" x 10" |
| Autoradiography film | GE Life Sciences | 28-9564-78 | Storage Phosphor Screen BAS-IP SR 2025 E Super Resolution, 20 x 25 cm, screen only |
| Butterfly Needle Catheter | SAI Infusion Technologies | BLF-24 | |
| DOTA-NHS-ester | Macrocyclics | B-280 | |
| EAE Induction Kit | Hooke Laboratories | EK-2160 | |
| Geiger Counter | Ludlum | 14C | |
| GNEXT PET/CT Scanner | Sofie | GNEXT | |
| Hidex Automatic Gamma Counter | Hidex | AMG | |
| HPLC Column | Phenomenex | 00H-2146-K0 | 5 μm SEC-s3000 400 Å, 300 x 7.8 mm |
| Illustra NAP-5 column | Cytiva | 17085301 | DNA gravity column |
| Image J | NIH | ARG analysis software | |
| Low Protein Binding Collection Tubes (1.5 mL) | Thermo Scientific | PI90410 | |
| NanoDrop Lite Spectrophotometer | Thermo Scientific | 840281400 | UV-Vis micro/nano-spectrophotometer |
| PCR tubes 0.2 mL, for DNA grade | Eppendorf | 30124707 | |
| Typhoon phosphor imager 9410 | GE Healthcare | 8149-30-9410 | |
| VivoQuant | Invicro | Version 4 Patch 3 | PET Analysis Software; must purchase brain atlas add-on |
| Zeba Spin Desalting Columns, 7K MWCO, 0.5 mL | Thermo Scientific | PI89882 | Desalting column |
Referências
- Dobson, R., Giovannoni, G. Multiple sclerosis – a review. European Journal of Neurology. 26 (1), 27-40 (2019).
- Karussis, D. The diagnosis of multiple sclerosis and the various related demyelinating syndromes: A critical review. Journal of Autoimmunity. 48-49, 134-142 (2014).
- Hauser, S. L., et al. B-cell depletion with Rituximab in relapsing-remitting multiple sclerosis. The New England Journal of Medicine. 358 (7), 676-688 (2008).
- Chen, D., Gallagher, S., Monson, N. L., Herbst, R., Wang, Y. Inebilizumab, a B cell-depleting anti-CD19 antibody for the treatment of autoimmune neurological diseases: Insights from preclinical studies. Journal of Clinical Medicine. 5 (12), 107 (2016).
- Stevens, M. Y., et al. Development of a CD19 PET tracer for detecting B cells in a mouse model of multiple sclerosis. Journal of Neuroinflammation. 17 (1), 275 (2020).
- Chaney, A. M., Johnson, E. M., Cropper, H. C., James, M. L. PET imaging of neuroinflammation using [11C]DPA-713 in a mouse model of ischemic stroke. Journal of Visualized Experiments. (136), e57243 (2018).
- Lyons, J. -. A., Ramsbottom, M. J., Cross, A. H. Critical role of antigen-specific antibody in experimental autoimmune encephalomyelitis induced by recombinant myelin oligodendrocyte glycoprotein. European Journal of Immunology. 32 (7), 1905-1913 (2002).
- Haugen, M., Frederiksen, J. L., Degn, M. B. cell follicle-like structures in multiple sclerosis-With focus on the role of B cell activating factor. Journal of Neuroimmunology. 273 (1-2), 1-7 (2014).
- James, M. L., et al. Imaging B cells in a mouse model of multiple sclerosis using 64Cu-Rituximab PET. Journal of Nuclear Medicine. 58 (11), 1845-1851 (2017).
- Zeglis, B. M., et al. An enzyme-mediated methodology for the site-specific radiolabeling of antibodies based on catalyst-free click chemistry. Bioconjugate Chemistry. 24 (6), 1057-1067 (2013).
- Lyons, J. -. A., et al. B cells are critical to induction of experimental allergic encephalomyelitis by protein but not by a short encephalitogenic peptide. European Journal of Immunology. 29 (11), 3432-3439 (1999).

