Isolation of Nuclei from Flash-Frozen Liver Tissue for Single-Cell Multiomics
Summary
Here, we present a protocol for isolating nuclei from flash-frozen, archived liver tissues for single-nucleus RNA-seq, ATAC-seq, and joint multiomics (RNA-seq and ATAC-seq).
Abstract
The liver is a complex and heterogenous tissue responsible for carrying out many critical physiological functions, such as the maintenance of energy homeostasis and the metabolism of xenobiotics, among others. These tasks are performed through tight coordination between hepatic parenchymal and non-parenchymal cells. Additionally, various metabolic activities are confined to specific areas of the hepatic lobule-a phenomenon called liver zonation. Recent advances in single-cell sequencing technologies have empowered researchers to investigate tissue heterogeneity at a single-cell resolution. In many complex tissues, including the liver, harsh enzymatic and/or mechanical dissociation protocols can negatively affect the viability or the quality of the single-cell suspensions needed to comprehensively characterize this organ in health and disease.
This paper describes a robust and reproducible protocol for isolating nuclei from frozen, archived liver tissues. This method yields high-quality nuclei that are compatible with downstream, single-cell omics approaches, including single-nucleus RNA-seq, assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq), as well as multimodal omics (joint RNA-seq and ATAC-seq). This method has been successfully used for the isolation of nuclei from healthy and diseased human, mouse, and non-human primate frozen liver samples. This approach allows the unbiased isolation of all the major cell types in the liver and, therefore, offers a robust methodology for studying the liver at the single-cell resolution.
Introduction
Single-cell genomics is rapidly becoming an essential methodology to study liver function and assess the impact of cellular heterogeneity in health and disease conditions1. The rapid development of "multiomics" for the simultaneous measurement of different layers of information and the parallel expansion of robust computational pipelines is paving the way for the discovery of previously unknown cell types and subtypes in the normal and diseased liver2.
The possibility of exploring biobanks and archived frozen samples has significantly increased the opportunities to revisit and discover the role of non-parenchymal cells3,4,5 and investigate the role of polyploid hepatocytes during aging and in chronic diseases6,7,8,9. Therefore, this paper describes a robust and reproducible single-nucleus isolation protocol for flash-frozen (FF) archived livers that is compatible with downstream single-nucleus RNA sequencing and ATAC sequencing, as well as with multimodal omics (joint RNA-seq and ATAC-seq) (Figure 1).
This workflow allows for the investigation of the transcriptome and the chromatin accessibility of all the cell types in the liver, independent of the cell size or fragility, in enzymatic dissociation protocols. It can be performed with small tissue sections (15-30 mg or 5-10 mm3) from precious human samples or transgenic mice. Determining the high purity of the nuclei isolation includes the quantification and measurement of the nuclear size, which might correlate with increased cell size and senescence10,11, and this purity is relevant for the analysis of both hepatocyte ploidy12 and cell size-dependent transcriptional mechanisms11,13,14,15. Additionally, nuclei isolated from frozen livers retain valuable information about liver zonation. The workflow and tissue collection allow for the validation of single-cell genomics data or further complementary analyses, such as immunohistochemistry or spatial transcriptomics from the same tissue and the same individual. Therefore, this approach can be applied to multiple liver disease conditions and model organisms systematically and reliably.
Protocol
All animal experiments were carried out in compliance with German Animal Welfare Legislation and the regulations of the Government of Upper Bavaria. Animal housing was approved according to §11 of the German Animal Welfare Act and performed in accordance with Directive 2010/63/EU.
1. Tissue preparation
- Sacrifice a 3 month old to 22 month old male C57BL/6J mouse by cervical dislocation. Lay the animal on a dissecting board, secure the extremities with pins, and disinfect the abdomen with 70% ethanol.
- Perform necropsy as recommended by Treuting et al.16.
- Open the abdomen up to the ribcage, visualize the liver, and use forceps to carefully remove the liver without piercing the lobules.
- Extract the intact liver by holding the diaphragm with forceps and removing the connecting tissue with scissors.
- Wash the organ in cold phosphate-buffered saline (PBS), pat it dry on a clean paper towel, and cut the liver lobules into several pieces for different purposes: FF for single-nucleus isolation and multiomics; paraformaldehyde-fixed (10% PFA) for paraffin embedding; and/or embedded in optimal cutting temperature (OCT) compound for further histological analyses (Figure 1A).
- Aliquot the liver pieces into cryovials or 5 mL screw-cap tubes, and immediately flash-freeze in liquid nitrogen. Store the frozen liver samples at −80 °C for downstream single-nucleus multiomics experiments (Figure 1B, C).
NOTE: The cryopreserved tissue can be safely stored at −80 °C for several years and should always be transported on dry ice at −80 °C prior to usage.
2. Nuclei isolation
- Benchtop cleaning and preparation of the buffers and consumables
- Clean the working benchtops and pipettes with 70% ethanol and RNase decontamination solution, or use dedicated RNase-free benchtops and materials.
- Prechill both the swinging bucket and fixed-angle centrifuges, 1.5 mL/2 mL tubes, and multi-well plates to 4 °C, as detailed below.
- Prechill the RNase-free, single-use tweezers used for tissue handling, a disposable sterile scalpel, and a Petri dish on dry ice (−60 °C).
- Prechill the Dounce glass homogenizer and pestles on ice (4 °C). Place each pestle (A and B) into a 5 mL tube to avoid direct contact with the ice and potential RNase contamination.
- Prepare a diluent solution of iodixanol medium (IDM) (Table 1A), and use it to make 50% and 29% dilutions of the 60% iodixanol stock density gradient medium (Table 1B and Table 1C, respectively).
- Prechill all the tubes. For each sample that is going to be processed, prepare the following tubes:
- Prepare three 1.5 mL low-DNA binding tubes (one for the filtered tissue homogenate, a second one containing 250 µL of a 50% dilution of iodixanol solution, and a third one for the clean nuclei suspension).
- Prepare one 2 mL round-bottom tube containing 500 µL of a 29% dilution of iodixanol solution for the density gradient separation.
- Prepare one 15 mL conical tube for the nuclei isolation medium-2 (NIM-2) and homogenization buffer (HB).
- Prepare the nuclei isolation medium-1 (NIM-1) (Table 1D), and use it to make the NIM-2 (Table 1E) and, subsequently, the HB (Table 1F). Add both the RNAse inhibitors just before use, as described below in the protocol.
- Prepare the nuclei storage buffer (NSB) as described in Table 1G. Add recombinant RNAse inhibitor just before use. Add protein-based RNAse inhibitor into the NSB before use for FACS sorting (optional).
- Prepare a 500 mL beaker with sterile water to soak the Dounce homogenizers and pestles following the tissue homogenization for the optimal cleaning and maintenance of the homogenizers.
- Tissue homogenization
- Cut a 20-30 mg (or 5 mm3) tissue piece with a prechilled scalpel inside the Petri dish on dry ice. Then, immediately transfer the Petri dish to wet ice (4 °C). Add 1 mL of HB, and using the cold scalpel, mince the tissue as much as possible to allow it to be easily aspirated with a 1 mL wide-orifice tip.
NOTE: Always use wide-orifice tips for all tissue/nuclei transfers. As an alternative, 1 mL tips can be cut with a sterile scalpel on a sterile plastic cover to generate wide orifices (Figure 2A). - Collect the tissue suspension, and transfer it to a prechilled 2 mL glass Dounce homogenizer (Figure 2B).
- Wash the Petri dish with an additional 0.5-1 mL of HB, and collect all the remaining tissue pieces while keeping everything on ice.
- Slowly and carefully do five strokes with loose pestle A on ice. Avoid creating bubbles by using twisting motions of the pestle when pulling it up and down. Ensure that the pestle moves carefully from the top to the bottom of the homogenizer with each stroke.
- Subsequently, perform 10-15 slow strokes with tight pestle B on ice. Avoid creating bubbles.
NOTE: It is recommended to visually inspect the nuclei under the microscope after 10 strokes with pestle B to ascertain if more strokes are needed. Do this by using trypan blue staining (1:1 ratio of trypan to sample) and a manual hemocytometer (e.g., mix 10 μL of trypan blue with 10 μL of nuclei suspension, and use 10 μL of the mixture for examination under the microscope; Figure 2C). - Filter the homogenate through a 50 µm cell strainer while transferring it into a prechilled 1.5 mL tube. Use more than one filter and/or tube for homogenates containing a high amount of connective tissue clumps.
- Rinse the homogenizer and the filters used with an additional 0.5-1 mL of HB to thoroughly collect all the tissue homogenate. Proceed to the density gradient centrifugation.
- Cut a 20-30 mg (or 5 mm3) tissue piece with a prechilled scalpel inside the Petri dish on dry ice. Then, immediately transfer the Petri dish to wet ice (4 °C). Add 1 mL of HB, and using the cold scalpel, mince the tissue as much as possible to allow it to be easily aspirated with a 1 mL wide-orifice tip.
- Density gradient centrifugation
- Centrifuge the filtered homogenate in a prechilled, fixed-angle centrifuge at 1,000 × g for 8 min at 4 °C.
- While the sample is spinning, prepare a 1.5 mL tube containing 250 µL of 50% iodixanol dilution and one 2 mL tube containing 500 µL of 29% iodixanol dilution. Keep both tubes on ice.
- Following centrifugation, aspirate the supernatant without disturbing the pellet using a vacuum pump.
NOTE: Using manual pipetting will compromise the quality of the final nuclei suspension. - Add 250 µL of HB to the pellet using the 1 mL wide-orifice pipette tip, and resuspend very slowly.
- Transfer 250 µL of the nuclei suspension into a prechilled 1.5 mL tube containing 250 µL of 50% iodixanol dilution, and mix gently but thoroughly to generate a 25% iodixanol/nuclei suspension.
- Transfer 500 µL of the 25% iodixanol/nuclei suspension into a prechilled 2 mL tube containing 500 µL of 29% iodixanol dilution.
NOTE: The 500 µL of the 25% iodixanol/nuclei suspension should be deposited gently on top of the 500 µL of 29% iodixanol solution such that the 25%/29% iodixanol mixtures show clear phase separation. Use the side of the tube wall with the pipette tip positioned at a 45° angle to create this gradient interface. From then on, the tube must be handled gently so as not to disturb this gradient. - Centrifuge the tube in a prechilled swinging bucket centrifuge at 12,500 g for 20 min with the brake set to OFF.
- Just before the centrifugation step is completed, add the RNAse inhibitors to the NSB buffer when proceeding to the scRNA-seq pipelines (see Table 1G).
- Following centrifugation, aspirate the supernatant without disturbing the pellet using a vacuum pump.
NOTE: Using manual pipetting will compromise the quality of the final nuclei suspension. - Using the 1 mL wide-orifice pipette tip, gently resuspend the pellet in 100-300 µL of NSB, and transfer the nuclei suspension into a clean, prechilled 1.5 mL tube.
- Count the nuclei using trypan blue solution (1:1 ratio of trypan to sample) and a manual hemocytometer (e.g., mix 10 μL of trypan blue with 10 μL of nuclei suspension, and use 10 μL of the mixture for counting; Figure 2D).
- Use the obtained nuclei suspension immediately for the single-nucleus genomics assay.
NOTE: The nuclei suspension in NSB can be stored refrigerated at 4 °C for up to 1 week for further analysis by flow and/or imaging flow cytometry but not for snRNA-seq or snATAC-seq.
3. Nuclei sorting for hepatocyte ploidy profiling or well-based sequencing approaches
- For flow cytometry-based cell sorting, filter the nuclei suspension through a 50 µm filter into a prechilled 5 mL FACS tube.
- Use a flow cytometry sorter fitted with a 100 μm nozzle. Load the FACS tube onto the sorter, and preview the sample.
- Set up the gating strategy for the nuclei sorting, starting with a scatter gate by plotting the forward scatter area versus the side scatter area (FSC-A/SSC-A), followed by Hoechst-Height versus Hoechst-Area (nuclei gate), and then Hoechst-Width versus Hoechst-Area (singlets gate). Visualize the nuclei ploidy profile on the Hoechst-Area histogram.
NOTE: For better peak resolution, visualize the Hoechst channel (450/50) on the linear scale (Figure 3A). - For sorting into 96-/384-well plates, set the droplet delay, and optimize the plate alignment using the colorimetric method with benzidine substrate-horseradish peroxidase (TMB-HRP), as described previously6.
- Set both the sample cooling and plate holder to chill at 4 °C, with the sample rotation turned on at 300 rpm.
- Sort the single nuclei at a sample concentration of ~1 x 105 nuclei/mL and with the flow rate at 200-500 events/s.
4. Visual inspection and quantification of nuclear parameters by imaging cytometry (optional)
- Load a 0.5 mL tube containing 50 µL of the nuclei suspension at a concentration of 2 × 107 nuclei/mL.
- Set up an all events gate based on the aspect ratio versus the Hoechst fluorescence channel to visualize the nuclei ploidy profile (Figure 3B).
- Acquire the sample at a 40x magnification using brightfield (BF) and the Hoechst fluorescence channel.
- Inspect and quantify 2n and 4n nuclei using the brightfield measurement and Hoechst fluorescence intensity with imaging cytometry software (Figure 3C).
5. Single-nucleus RNA-seq, ATAC-seq, or multiome library construction and sequencing
- For droplet-based snRNA-seq approaches, load the purified nuclei suspension directly into the microfluidic device for automated parallel partitioning and molecular barcoding17.
- After the microfluidic run is completed in the single-cell partitioning device, collect the gel bead-encapsulated nuclei, incubate, and clean as described previously in the manufacturer's guidelines17.
- Perform 11 polymerase chain reaction (PCR) cycles for the cDNA preamplification using the following program: 3 min at 98 °C, (15 s at 98 °C, 20 s at 63 °C, and 1 min at 72 °C) x 11, 1 min at 72 °C, and hold at 4 °C. Continue with an end repair and A-tailing step and adaptor ligation, as indicated by the manufacturer17. For the subsequent final gene expression library construction, perform 10 PCR cycles using the following program: 45 s at 98 °C, (20 s at 98 °C, 30 s at 54 °C, 20 s at 72 °C) x 10, 1 min at 72 °C, and hold at 4 °C.
- Sequence the obtained libraries to a read depth of ~20,000-50,000 mean reads per nucleus.
- For the joint multiomics (RNA + ATAC) droplet-based sequencing, incubate the liver nuclei in lysis buffer for 5 min, and then tagment them for 1 h, as described previously18.
- Load the tagmented nuclei directly into a microfluidic device for automated parallel partitioning and molecular barcoding.
- After the microfluidic run is completed, collect the gel bead-encapsulated nuclei, incubate, and clean as described by the manufacturer18.
- Perform six PCR cycles for the cDNA preamplification step using the following program: 5 min at 72 °C, 3 min at 98 °C, (20 s at 98 °C, 30 s at 63 °C, 1 min at 72 °C) x 6, 1 min at 72 °C, and hold at 4 °C.
- Take 35 µL of the preamplified sample, and perform a cDNA amplification as follows: 3 min at 98 °C, (15 s at 98 °C, 20 s at 63 °C, 1 min at 72 °C) x 6, 1 min at 72 °C, and hold at 4 °C. Continue with the end repair and A-tailing step and adaptor ligation, as indicated by the manufacturer18 . For the subsequent final sample indexing PCR, perform 15 PCR cycles as follows: 45 s at 98 °C, (20 s at 98 °C, 30 s at 54 °C, 20 s at 72 °C) x 15, 1 min at 72 °C, and hold at 4 °C.
- For the construction of ATAC libraries, use 40 µL, and amplify for six PCR cycles for sample indexing using the following program: 45 s at 98 °C, (20 s at 98 °C, 30 s at 67 °C, 20 s at 72 °C) x 6, 1 min at 72 °C, and hold at 4 °C.
- Sequence the obtained multiome gene expression libraries to a minimum read depth of 20,000 read pairs per nucleus and the multiome ATAC libraries to a minimum read depth of 25,000 read pairs per cell, as recommended by the manufacturer18.
Representative Results
This workflow for single-nucleus isolation from frozen liver samples is tailored for single-nucleus multiomics and relies on three main steps, which can be summarized as i) sample collection for the parallel analysis of cellular heterogeneity and tissue architecture, ii) single-nucleus suspension, and iii) single-nucleus multiomics (Figure 1). The extracted livers are dissected from euthanized mice and cut into pieces for histological inspection for either paraffin-embedding, cryosectioning, or both. Other cut pieces are immediately flash-frozen in liquid nitrogen for downstream, single-nucleus isolation for multiomics analyses. This system of tissue collection allows the user to further validate the single-nucleus omics data on tissue sections from the same individual, thereby complementing the data set with spatial transcriptomics or with immunohistochemistry analyses, if needed.
The microscopic examination of the nuclei extracted from frozen livers with the method described here shows that the density gradient centrifugation step greatly facilitates the removal of unwanted cellular and tissue debris (Figure 2C,D). Furthermore, this methodology preserves all levels of ploidy, which can be validated and quantified by cytometric analyses (Figure 3).
To further validate the performance of this protocol, we conducted droplet-based snRNA-seq on unsorted and FACS-sorted nuclei and analyzed the data following the Seurat pipeline. In brief, the extracted single nuclei were prepared for droplet-based snRNA-seq as described previously19. For the snRNA-seq of 2n and 4n nuclei or higher levels of ploidy, 11 cycles were used for the cDNA amplification step and 10 cycles for the final gene expression library construction. The resulting libraries were sequenced to a read depth of ~25,000-39,000 mean reads per nucleus. The obtained single nuclei reads were mapped to the GRCm39/mm39 mouse genome. When running the preprocessing pipeline, the command –include-intron was added to ascertain the inclusion and quantification of the unspliced messenger RNA (mRNA) present in the nuclei. The EmptyDrops algorithm incorporated within the aligner filtered out and removed the empty droplets.
The R package Seurat (version 4.1.1) was used to compute quality control (QC) metrics using the unique molecular identifier (UMI) count matrix as outputted by the snRNA-seq analysis pipeline. Counts with less than 100 features (genes) and less than 10 cells were removed. The nuclei were filtered according to identified QC thresholds: minimum number of genes = 200 and maximum number of genes = 8,000, mitochondrial fraction <1% and ribosomal fraction <2%. The top 3,000 highly variable genes (HVGs) were used for the principal component analysis (PCA), as implemented in Seurat. Graph-based clustering was performed after inputting the top 15 PC dimensions resulting from the PCA analysis. To cluster the cells, we applied the modularity optimization technique (Louvain algorithm) with a resolution parameter set to 0.5. To visualize and explore this dataset, we ran non-linear dimensional reduction, namely Uniform Manifold Approximation and Projection (UMAP). The identity of each cluster was assigned based on prior knowledge of the marker genes6,20,21.
Figure 4A shows high-quality metrics from the data obtained using this method of nuclei extraction. UMAP represents the number of counts across all the nuclei and the major cell types that can be confidently identified only with the nuclear transcriptome and relatively shallow sequencing (~25,000-40,000 mean reads per cell) (Figure 4B). This approach permits the investigation of liver-specific transcription factors such as Hnf4a, Mlxipl, and Ppara, as well as downstream target genes involved in the metabolism of xenobiotics (i.e., Cyp2e1 and Cyp2f2)(Figure 4C). Of note, the extracted nuclei retained critical information about liver zonation, as shown by the complementary pattern of hallmark genes such as the pericentral Cyp2e1 and the periportal Cyp2f2 (Figure 4C).
We further assessed the compatibility of the unsorted nuclei extracted using this method with newer multiomics assays for the simultaneous profiling of the epigenomic landscape (ATAC) and gene expression (RNA) in the same single nuclei18. We optimized the nuclei lysis incubation time to 5 min prior to transposition. The sequencing libraries were constructed by running six PCR cycles for the cDNA preamplification. From the preamplified sample, 35 µL was taken and further amplified by 15 PCR cycles for sample indexing to construct the gene expression library. A representative electropherogram of the cDNA trace is shown in Figure 5A (top). For the construction of the ATAC libraries, 40 µL of preamplified sample was used and run for an additional six PCR cycles for sample indexing, with the representative electropherogram of the DNA trace shown in Figure 5A (bottom). The resulting gene expression library (RNA) was sequenced to a depth of 44,600 reads per nucleus and the ATAC library to a depth of 43,500 reads per nucleus (Figure 5B).
Similar to the above-described, single-modality, droplet-based sequencing protocol, read mapping, alignment, empty drop removal, and fragment counting were performed following standard guidelines, as described previously, using the GRCm39/mm39 reference genome18. Using the Seurat and Signac packages22, we performed a "weighted nearest neighbor" (WNN) analysis for multiple measurements of both modalities (RNA + ATAC) (Figure 5C), which allowed us to identify and annotate both the major and minor liver cell types without prominent biases due to cell size or nuclear fragility (Figure 5D). The pipeline, as published by the Satija laboratory22,23, includes standard QC steps-preprocessing and dimensional reduction-done on both assays independently. To have a good representation of the weighted combination of the RNA-seq and ATAC-seq modalities, the WNN graph was plotted and used for UMAP visualization, clustering, and annotation based on previously identified marker genes6,20,21. Similar to the assays using single modality, we also detected upstream transcriptional regulators (Hnf4a, Ppara, Mlxipl) and hallmark genes of liver zonation (Hamp, Cyp2e1, and Cyp2f2) (Figure 5E).
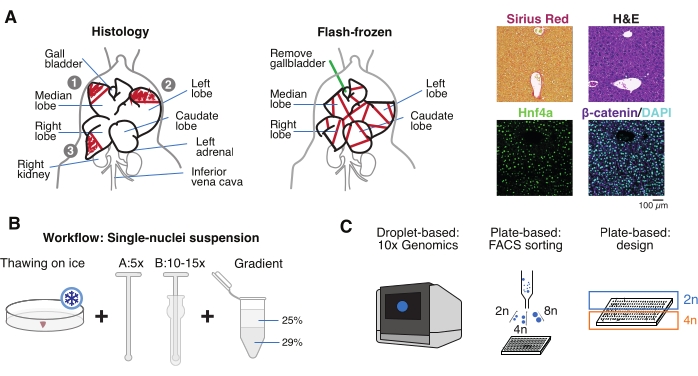
Figure 1: Experimental overview, workflow, and single-cell genomic applications. (A) Illustrative representation of tissue sampling for histology (left, three sections are selected for paraffin embedding and/or cryosectioning), flash-frozen tissue collection for single-cell genomics (middle), and representative immunohistochemistry and immunofluorescence analyses (right); scale bar = 100 µm. (B) Critical steps for high-quality single-nucleus suspensions. (C) The nuclei suspensions can be loaded onto a 10x chromium chip or used for FACS sorting and plate-based approaches. Abbreviations: H&E = hematoxylin and eosin; DAPI = 4',6-diamidino-2-phenylindole; FACS = fluorescence-activated cell sorting. Please click here to view a larger version of this figure.
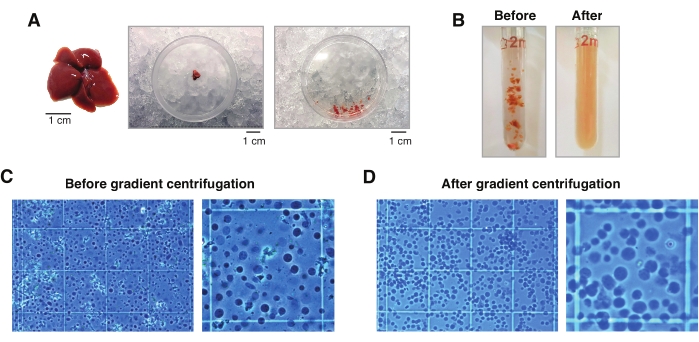
Figure 2: Liver dissection and Dounce-glass tissue homogenization. (A) Representative murine liver from a 3 month old C57BL6/J mouse (left); the liver section used for the single-nucleus isolations before (middle) and after mincing the tissue with the scalpel (right). Scale bars = 1 cm. (B) Illustrative images of 2 mL Dounce-glass homogenization before strokes with "loose" pestle A (left) and after "tight" pestle B (right). Monitoring the tissue homogenization using a hemocytometer (C) before gradient centrifugation and (D) after gradient centrifugation. Please click here to view a larger version of this figure.
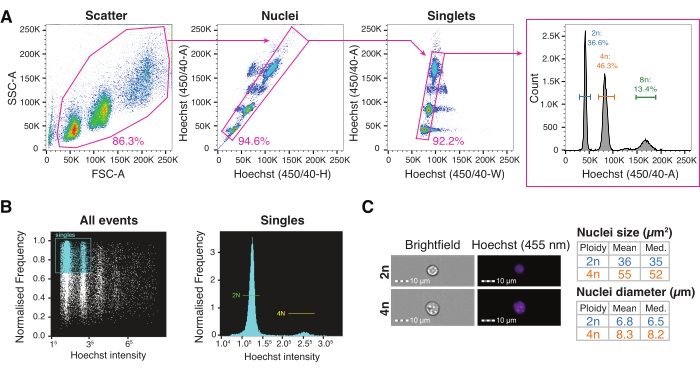
Figure 3: Fluorescence-activated cell sorting and imaging flow cytometry for high-throughput nuclei characterization. (A) Gating strategy for single-nucleus FACS sorting into a plate for the interrogation of different levels of ploidy. Scatter gate based on the forward scatter area versus the side scatter area set to exclude debris; nuclei gate based on Hoechst-Height versus Hoechst-Area incorporating multiple nuclei populations; singlets gate based on Hoechst-Width versus Hoechst-A set for doublet discrimination; the Hoechst-A histogram allows the visualization of the nuclei ploidy profile. (B) Representative imaging cytometry quantification of all the events (left) and single (right) events, showing diploid and tetraploid nuclei. (C) Brightfield and Hoechst images of 2n and 4n nuclei and their quantification using imaging cytometry. Abbreviations: FACS = fluorescence-activated cell sorting; FSC-A = forward scatter area; SSC-A = side scatter area; Hoechst-H = Hoechst-Height; Hoechst-A = Hoechst-Area; Hoechst-W = Hoechst-Width. Please click here to view a larger version of this figure.
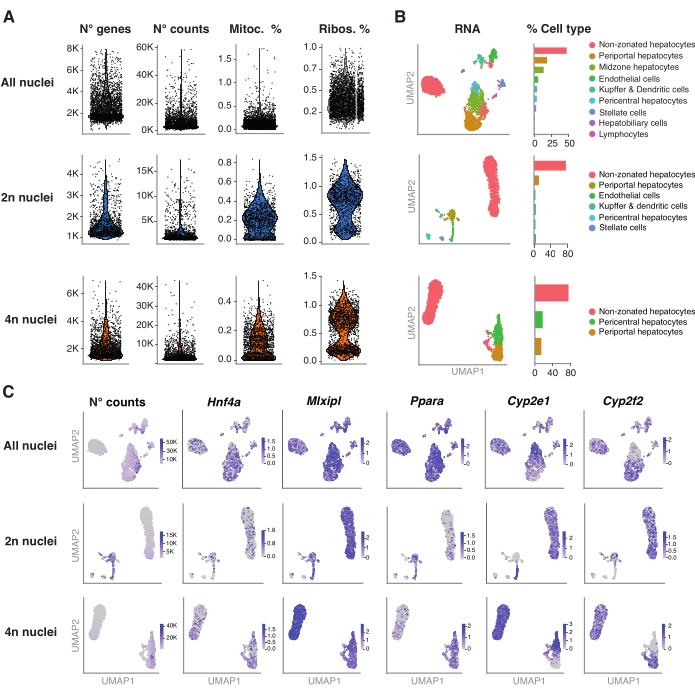
Figure 4: Deep characterization of hepatocyte ploidy with snRNA-seq. (A) Violin plots showing the number of genes, counts, percentage of mitochondrial genes, and percentage of ribosomal genes detected. (B) UMAP demonstrating the cell types detected using snRNA-seq (left), with the quantification expressed in percentage of nuclei (right). (C) UMAP illustrates the number of counts and indicated the expression of hepatocyte-specific genes. All the nuclei (top row), 2n nuclei (middle row), and 4n nuclei (bottom row). Abbreviations: snRNA-seq = single-nucleus RNA-seq; UMAP = Uniform Manifold Approximation and Projection; mitoc. = mitochondrial genes; Ribos. = ribosomal genes. Please click here to view a larger version of this figure.
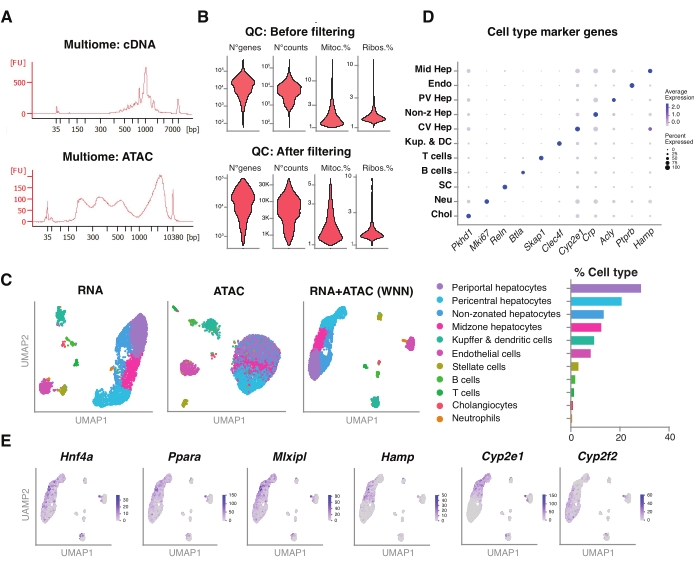
Figure 5: Quality control and analysis of multiomics (joint RNA-seq and ATAC-seq) from frozen, archived young livers. (A) Representative automatic electrophoretic traces obtained after the multiomic pipeline showing the molecular weight product after cDNA synthesis and ATAC. (B) Violin plot demonstrating the number of counts for ATAC-seq, RNA-seq, the percentage of mitochondrial genes, and the percentage of ribosomal genes before and after filtering. (C) UMAP shows the gene expression from RNA-seq (left), ATAC-seq (middle), and joint modalities-RNA-seq and ATAC-seq (right). (D) Different cell types are annotated in different colors, with the expression of indicated hepatocyte-specific genes. (E) Feature plot showing the cluster-specific expression of the indicated genes in the indicated cell types. Abbreviations: ATAC-seq = assay for transposase-accessible chromatin with high-throughput sequencing; Mid Hep = mid-zonal hepatocytes; Endo = endothelial cells; PV Hep = periportal hepatocytes; Non-z Hep = non-zonated hepatocytes; CV Hep = pericentral hepatocytes; Kup & DC = Kupffer & dendritic cells; SC = stellate cells, Neu = neutrophils; Chol = cholangiocytes; mitoc. = mitochondrial genes; Ribos. = ribosomal genes. Please click here to view a larger version of this figure.
| Reagent | Stock | 10 mL | 15 mL | 50 mL | ||
| (A) Iodixanol Medium (IDM) | ||||||
| 250 mM Sucrose | 1M | 12.5 mL | ||||
| 150 mM KCl | 2M | 3.75 mL | ||||
| 30 mM MgCl2 | 1M | 1.5 mL | ||||
| 60 mM Tris buffer pH 8.0 | 1M | 3 mL | ||||
| Ultrapure RNase free water | 29.25 mL | |||||
| (B) 50 % IDM | ||||||
| Iodixanol | 60% | 12.5 mL | ||||
| IDM | 2.5 mL | |||||
| (C) 29% IDM | ||||||
| Iodixanol | 60% | 7.25 mL | ||||
| IDM | 7.75 mL | |||||
| (D) Nuclei Isolation Medium-1 (NIM-1) | ||||||
| 250 mM Sucrose | 1M | 12.5 mL | ||||
| 25 mM KCl | 2M | 0.625 mL | ||||
| 5 mM MgCl2 | 1M | 0.25 mL | ||||
| 10 mM Tris buffer pH 8.0 | 1M | 0.5 mL | ||||
| Ultrapure Rnase-free water | 36.125 mL | |||||
| (E) Nuclei Isolation Medium-2 (NIM-2) | ||||||
| NIM-1 buffer | 9.99 mL | |||||
| Dithiothreitol (DTT) | 1 mM | 0.01 mL | ||||
| Protease Inhibitor tablet (EDTA-free) | 1 | 1 Tablet | ||||
| (F) Homogenization Buffer (HB) | ||||||
| NIM-2 buffer | 9.697 mL | |||||
| Recombinant RNase Inhibitor | 40 U/µL | 0.1 mL | ||||
| Protein-based RNAse Inhibitor (SUPERase•IN) | 20 U/µL | 0.1 mL | ||||
| 0.1% Triton-X | 10% | 0.1 mL | ||||
| 3 µg/mL Hoechst 33342 | 10 mg/mL | 0.003 mL | ||||
| (G) Nuclei Storage Buffer (NSB) | ||||||
| 166.5 mM Sucrose | 1M | 1.665 mL | ||||
| 5 mM MgCl2 | 1M | 0.05 mL | ||||
| 10 mM Tris pH 8.0 | 1M | 0.1 mL | ||||
| Recombinant RNase Inhibitor | 40 U/µL | 0.1 mL | ||||
| Protein-based RNase Inhibitor (SUPERase•IN) * | 20 U/µL | 0.1 mL | ||||
| Ultrapure RNase free water | 8.085 mL | |||||
| * Optional (for FACS sorting only) | ||||||
Table 1: Solution recipes. (A) Preparation of iodixanol medium (IDM); (B) 50% dilution of iodixanol solution; (C) 29% dilution of iodixanol solution. (D) Preparation of nuclei isolation medium-1 (NIM-1). (E) Preparation of nuclei isolation medium-2 (NIM-2). (F) Preparation of homogenization buffer (HB). (G) Preparation of nuclei storage buffer (NSB).
Discussion
Dissecting the cellular composition of the liver by single-cell or single-nucleus RNA-seq provides a deeper understanding of liver disease development and progression3,4,5,24. Single-cell isolation from livers is time-consuming and requires protocols that involve harsh mechanical or enzymatic dissociation25,26,27. It is widely accepted that every tissue requires a systematic evaluation to determine the optimal tissue dissociation protocol, as well as a suitable storage method to capture fragile cell types or nuclei28. Depending on the tissue availability, disease of interest, developmental stage, or model organism, the preparation of a single-nucleus suspension for downstream processing might be a more suitable methodology than using single-cell suspensions. Importantly, in the liver, scRNA-seq and snRNA-seq have shown a high correlation between nuclear and cytoplasmic mRNA, suggesting that both approaches present complementary information2,3,4,6,29.
This paper provides a standardized, robust, and reproducible single-nucleus isolation from frozen, archived liver samples from mice and other species, including humans and macaques. This method can be used for wild-type mice fed with chow and a high-fat diet (HFD) and for mouse models of liver fibrosis using both well plate-based and droplet-based single-nucleus genomic approaches6. This method relies on the protocol described originally for brain tissue by Krishnaswami et al.30 with additional modifications tailored for flash-frozen liver. Optimal homogenization liberates most of the nuclei from the tissue without negatively affecting the nuclear membrane integrity. Overdouncing, however, can damage the fragile nuclei and decrease their overall quality. Young and/or fatty livers usually require only 5 strokes with pestle A and 10 strokes with pestle B, while old and/or fibrotic livers might require 15 strokes with pestle B but not more. It is not recommended, therefore, to perform more strokes beyond the numbers indicated here. Overdouncing might negatively affect the quality of the single-nucleus suspension and increase the amount of ambient RNA. Subsequently, this might lead to the need for performing additional computational filtering steps during the downstream data analyses.
The protocol presented here is versatile and can be adjusted to different liver conditions in young (3 months) and old (24 months) mice. Since we found that a larger section of the liver is necessary for old, HFD, and fibrotic tissues, the size of the tissue available for processing can pose a limitation for some users with smaller amounts of starting biological material. However, gradient purification is highly recommended for immediate sample processing with droplet-based genomic assays. If the nuclei need to be FACS sorted into 96-/384-well plates for well-based assays, gradient purification can be omitted. We do encourage users still to perform the gradient purification if there is enough tissue sample to obtain the recommended nuclei concentration for FACS sorting (i.e., ~1 × 105 nuclei/mL).
The liver is characterized by the polyploid nature of hepatocytes9, but the role of hepatocyte ploidy in normal physiology and disease is not yet clear. There is a growing body of evidence indicating that ploidy provides genomic variability31, and it is well known that ploidy increases with age32,33. The enrichment of mononucleated tetraploid hepatocytes, however, is also clinically associated with poor prognosis in human hepatocellular carcinoma (HCC)34. Similarly, changes in hepatocyte ploidy levels are linked to aging-related chronic liver diseases such as non-alcoholic fatty liver disease (NAFLD)35,36,37. Ploidy is the condition of possessing more than two copies of the genome, which can be explored by staining the genome content with a DNA dye such as Hoechst38. Hoechst dye, which is added to the HB prior to extraction, labels all the nuclei during the isolation protocol. This allows for the distinction between diploid and polyploid nuclei based on their DNA content when excited by a UV (350 nm) or violet (450 nm) laser on a flow cytometry instrument. With the showcased gating strategy, 2n, 4n, 8n, and higher levels of hepatocyte ploidy can be investigated in frozen, archived livers to better understand the role of cellular heterogeneity in tissue function1 (Figure 3A). Furthermore, the nuclear morphology, including the size and volume, can be quantified using imaging flow cytometry to correlate changes in the nucleus size with changes in the total number of counts or number of genes depending on the ploidy level (Figure 3B,C).
Multimodal omics measurement offers the opportunity to investigate several layers of genomic organization simultaneously. The joint RNA + ATAC multiomics approach allows the investigation of upstream regulators and downstream metabolic genes, providing a comprehensive approach for studying transcriptional networks and the chromatin architecture associated with liver function at the single-cell resolution. Furthermore, with the advances in computational methods that can account for data sparsity and the reduction in sequencing costs, single-cell multiomics is pioneering the assessment of multiple modalities from the same cell. This single-nucleus isolation protocol is compatible with the individual and joint assessment of expression and chromatin data sets. We have used standard pipelines established by Stuart et al.23 (Signac package) to illustrate the quality of the data, whilst several available and alternative computational methods can be easily adopted for downstream analyses23,39,40,41.
Overall, single-nucleus multiomics allows the investigation of bio-archived, FF mouse, human, and non-human primate liver tissues using a very small amount of starting sample material by implementing the nucleus-extraction protocol presented here. This invaluable tool will empower liver biologists to interrogate both gene expression and chromatin accessibility in the context of various liver pathologies. Additionally, various levels of hepatocyte ploidy and the resulting adjustment of gene expression dependent on their location in the liver lobule might reveal their role in liver pathologies. Therefore, we anticipate that the investigation of cellular heterogeneity will provide new opportunities for the development of precision medicine and targeted interventions against diseases such as HCC and NAFLD.
Declarações
The authors have nothing to disclose.
Acknowledgements
This research was supported by the Helmholtz Pioneer Campus (M.S., K.Y., C.P.M.-J.) and the Institute of Computational Biology (C. T.-L.). This research was also supported by AMED under Grant Number JP20jm0610035 (C.P.M.-J.). We thank the Core Genomics at HMGU (I. de la Rosa) and Bioinformatics (T. Walzthoeni) support, in particular Xavier Pastor for training and guidance. We thank A. Feuchtinger, U. Buchholz, J. Bushe, and all other staff members from the HMGU Pathology and Tissue Analytic core facility for their technical and scientific support, as well as, J. Zorn, R. Erdelen, D. Würzinger, staff members of E-Streifen, as well as the the Laboratory Animal Services core facility for their on-going scientific support and discussion. We are grateful to the Core Facility Cell Analysis at TranslaTUM (R. Mishra), and Luminex, A DiaSorin Company (P. Rein). We thank Dr. I Deligiannis for his technical support. Dr. M. Hartman, Dr. A. Schröder, and Ms. A. Barden (Helmholtz Pioneer Campus) were fundamental for their legal, managerial, and administrative support.
Materials
| 10% Tween 20 – 5 mL | Bio-Rad | 1662404 | |
| 2100 Bioanalyzer Instrument | Agilent | G2939BA | |
| Adhesive PCR film | Thermo Fisher Scientific | AB0558 | |
| Bioanalyzer High Sensitivity DNA Analysis | Agilent | 5067-4626 | |
| C1000 Touch Thermal Cycler | Bio-Rad | 1851196 | |
| Cell Sorter | For fluoresence-activated cell sorting (FACS); e.g. BD FACSAria Cell Sorter. | ||
| Centrifuge 5430R | Eppendorf | 5428000619 | Use chilled at 4 °C |
| Chromium Controller & Next GEM Accessory Kit | 10X Genomics | 1000204 | |
| Chromium Next GEM Chip G Single Cell Kit | 10X Genomics | 1000127 | |
| Chromium Next GEM Chip J Single cell kit, 16 reactions | 10X Genomics | 1000230 | |
| Chromium Next GEM Single Cell 3' Kit v3.1, 4 reactions | 10X Genomics | 1000269 | |
| Chromium Next GEM Single Cell Multiome ATAC + Gene Expression Reagent Bundle, 4 reactions | 10X Genomics | 1000285 | |
| cOmplete, EDTA-free Protease Inhibitor Cocktail | Sigma Aldrich | 11873580001 | |
| Dithiothreitol (DTT) 1 M solution | Sigma Aldrich | 646563 | Make 1 mM stock solution and use at 1 µM final concentration. |
| DNA AWAY Surface Decontaminant | Thermo Fisher Scientific | 10223471 | Wipe surfaces and pipettes before start of experiment |
| DNA LoBind Tubes, 1.5 mL | Thermo Fisher Scientific | 16628742 | |
| DNA LoBind Tubes, 2.0 mL | Thermo Fisher Scientific | 16638742 | |
| Elution Buffer (EB) – 250 mL | Qiagen | 19086 | |
| Eppendorf ThermoMixer C | Thermo Fisher Scientific | 13527550 | |
| Eppendorf ThermoMixer C Accessory: Smartblock | Thermo Fisher Scientific | 13518470 | |
| Ethanol, Absolute (200 Proof), Molecular Biology Grade – 100 mL | Thermo Fisher Scientific | 10517694 | |
| Filters 50 µm, sterile | SYSMEX PARTEC – CELLTRICS | 04-004-2327 | Adjust filter diameter according with tissue and nuclei size |
| Glycerin (Glycerol), 50% (v/v) – 1 L | Ricca Chemical Company | 3290-32 | |
| Hard-Shell, 384-Well PCR Plates, thin wall, skirted, clear/white | Bio-Rad | HSP3905 | |
| Herenz Heinz ABS Forceps | Thermo Fisher Scientific | 1131884 | |
| Hoechst 33342, Trihydrochloride, Trihydrate, 10 mg/mL Solution in Water | Invitrogen | H3570 | Light-sensitive |
| Imaging Flow Cytometer | For imaging flow cytometry analysis, e.g. Luminex Amnis ImageStream | ||
| Invitrogen TE Buffer – 100 mL | Thermo Fisher Scientific | 11568846 | |
| KCl (2 M), RNase-free, 100 mL | Thermo Fisher Scientific | AM9640G | |
| MgCl2 (1 M), 100 mL | Thermo Fisher Scientific | AM9530G | |
| MicroAmp 8-Cap Strip, clear-300 strips | Thermo Fisher Scientific | 10209104 | |
| MicroAmp 8-Tube Strip, 0.2 mL-125 strips | Thermo Fisher Scientific | 10733087 | |
| Mörser 2 mL DOUNCE | Wagner & Munz GmbH | 9651632 | RNase zap and rinse with MillQ before use |
| MyFuge 12 Mini MicroCentrifuge C1012 | Benchmark Scientific | C1012 | Or any other strip and tube mini centrifuge |
| Neubauer Hemocytometer | OMNILAB LABORZENTRUM | 5435293 | Visualize and count nuclei under microscope |
| Nuclease-Free Water (not DEPC-Treated) | Thermo Fisher Scientific | AM9937 | |
| OptiPrep Density Gradient Medium | Sigma Aldrich | D1556 | |
| Pipette tips RT LTS 1000 µL, Wide-O | Mettler Toledo | 30389218 | |
| Pistill "A" 2 mL | Wagner & Munz GmbH | 9651621 | RNase zap and rinse with MillQ before use |
| Pistill "B" 2 mL | Wagner & Munz GmbH | 9651627 | RNase zap and rinse with MillQ before use |
| Polypropylene 15 mL Centrifuge Tube | Thermo Fisher Scientific | 10579691 | |
| Polystyrene Petri dish, 60 mm x 15 mm | Thermo Fisher Scientific | 10634141 | |
| Polystyrene Round-Bottom 5 mL FACS Tubes | Thermo Fisher Scientific | 10100151 | |
| Protector RNase inhibitor – 2,000 U | Sigma Aldrich | 3335399001 | Keep in -20 °C until use |
| Protein-based RNase Inhibitor SUPERase•In (20 U/μL) | Thermo Fisher Scientific | AM2696 | Keep in -20 °C until use |
| Recombinant RNase Inhibitor | Clontech Takara | 2313B | Keep in -20 °C until use |
| RNAse free microfuge tubes – 0.5 mL | Thermo Fisher Scientific | AM12450 | |
| RNaseZap RNase Decontamination Solution | Thermo Fisher Scientific | AM9780 | Wipe surfaces and pipettes before start of experiment |
| SPRIselect – 60 mL | Beckman Coulter | B23318 | Aliquot and store in 4 °C |
| Sucrose, 500 g | Sigma Aldrich | S0389-500G | Make a 1 M stock solution |
| Swann-Morton Sterile Disposable Stainless Steel Scalpels | Thermo Fisher Scientific | 11798343 | |
| Tris-HCI (1M), pH 8.0 | Invitrogen | 15568025 | |
| Triton X-100, 98%, 100 mL | Thermo Fisher Scientific | 10671652 | Make 10% stock solution. Keep at 4 °C and protect from light. |
| Trypan Blue Solution, 0.4% | Thermo Fisher Scientific | 11538886 | |
| Vortex- Mixer | VWR | 444-1372 | Or any other type of vortex |
Referências
- Kamies, R., Martinez-Jimenez, C. P. Advances of single-cell genomics and epigenomics in human disease: where are we now. Mamm Genome. 31 (5-6), 170-180 (2020).
- Ramachandran, P., Matchett, K. P., Dobie, R., Wilson-Kanamori, J. R., Henderson, N. C. Single-cell technologies in hepatology: New insights into liver biology and disease pathogenesis. Nature Reviews Gastroenterology & Hepatology. 17 (8), 457-472 (2020).
- Ramachandran, P., et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 575 (7783), 512-518 (2019).
- Andrews, T. S., et al. Single-cell, single-nucleus, and spatial RNA sequencing of the human liver identifies cholangiocyte and mesenchymal heterogeneity. Hepatology Communications. 6 (4), 821-840 (2022).
- Xiong, X., et al. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Molecular Cell. 75 (3), 644-660 (2019).
- Richter, M. L., et al. Single-nucleus RNA-seq2 reveals functional crosstalk between liver zonation and ploidy. Nature Communications. 12 (1), 4264 (2021).
- Matsumoto, T., Wakefield, L., Tarlow, B. D., Grompe, M. In vivo lineage tracing of polyploid hepatocytes reveals extensive proliferation during liver regeneration. Cell Stem Cell. 26 (1), 34-47 (2020).
- Chen, F., et al. Broad distribution of hepatocyte proliferation in liver homeostasis and regeneration. Cell Stem Cell. 26 (1), 27-33 (2020).
- Donne, R., Saroul-Ainama, M., Cordier, P., Celton-Morizur, S., Desdouets, C. Polyploidy in liver development, homeostasis and disease. Nature Reviews. Gastroenterology & Hepatology. 17 (7), 391-405 (2020).
- Lengefeld, J., et al. Cell size is a determinant of stem cell potential during aging. Science Advances. 7 (46), 0271 (2021).
- Lanz, M. C., et al. Increasing cell size remodels the proteome and promotes senescence. Mol Cell. 82 (17), 3255-3269 (2022).
- Kim, J. Y., et al. PIDDosome-SCAP crosstalk controls high-fructose-diet-dependent transition from simple steatosis to steatohepatitis. Cell Metabolism. 34 (10), 1548-1560 (2022).
- Padovan-Merhar, O., et al. Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms. Molecular Cell. 58 (2), 339-352 (2015).
- Miettinen, T. P., et al. Identification of transcriptional and metabolic programs related to mammalian cell size. Current Biology. 24 (6), 598-608 (2014).
- Vargas-Garcia, C. A., Ghusinga, K. R., Singh, A. Cell size control and gene expression homeostasis in single-cells. Current Opinion in Systems Biology. 8, 109-116 (2018).
- Knoblaugh, S. E., Randolph-Habecker, J. . Necropsy and histology. In Comparative Anatomy and Histology: A Mouse, Rat, and Human Atlas (Second Edition). , (2018).
- Chromium Next GEM Single Cell 3 Reagent Kits v3.1 User Guide. Document number CG000204 (Rev D). 10x Genomics Available from: https://www.10xgenomics.com/support/single-cell-gene-expression/documentation/steps/library-prep/chromium-single-cell-3-reagent-kits-user-guide-v-3-1-chemistry (2019)
- Chromium Next GEM Single Cell Multiome ATAC + Gene Expression User Guide. Document number CG000338 (Rev D). 10x Genomics Available from: https://www.10xgenomics.com/support/single-cell-multiome-atac-plus-gene-expression/documentation/steps/library-prep/chromium-next-gem-single-cell-multiome-atac-plus-gene-expression-reagent-kits-user-guide (2021)
- MacParland, S. A., et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nature Communications. 9 (1), 4383 (2018).
- Martinez-Jimenez, C. P., Kyrmizi, I., Cardot, P., Gonzalez, F. J., Talianidis, I. Hepatocyte nuclear factor 4alpha coordinates a transcription factor network regulating hepatic fatty acid metabolism. Molecular and Cell Biology. 30 (3), 565-577 (2010).
- Schmidt, D., et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 328 (5981), 1036-1040 (2010).
- Hao, Y., et al. Integrated analysis of multimodal single-cell data. Cell. 184 (13), 3573-3587 (2021).
- Stuart, T., Srivastava, A., Madad, S., Lareau, C. A., Satija, R. Single-cell chromatin state analysis with Signac. Nature Methods. 18 (11), 1333-1341 (2021).
- Sampaziotis, F., et al. Cholangiocyte organoids can repair bile ducts after transplantation in the human liver. Science. 371 (6531), 839-846 (2021).
- Gomez-Lechon, M. J., Donato, M. T., Castell, J. V., Jover, R. Human hepatocytes as a tool for studying toxicity and drug metabolism. Current Drug Metabolism. 4 (4), 292-312 (2003).
- Gomez-Lechon, M. J., Donato, M. T., Castell, J. V., Jover, R. Human hepatocytes in primary culture: The choice to investigate drug metabolism in man. Current Drug Metabolism. 5 (5), 443-462 (2004).
- vanden Brink, S. C., et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nature Methods. 14 (10), 935-936 (2017).
- Denisenko, E., et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biology. 21 (1), 130 (2020).
- Nault, R., Fader, K. A., Bhattacharya, S., Zacharewski, T. R. Single-nuclei RNA sequencing assessment of the hepatic effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin. Cellular and Molecular Gastroenterology and Hepatology. 11 (1), 147-159 (2021).
- Krishnaswami, S. R., et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nature Protocols. 11 (3), 499-524 (2016).
- Duncan, A. W., et al. Aneuploidy as a mechanism for stress-induced liver adaptation. Journal of Clinical Investigation. 122 (9), 3307-3315 (2012).
- Kreutz, C., et al. Hepatocyte ploidy is a diversity factor for liver homeostasis. Frontiers in Physiology. 8, 862 (2017).
- Hunt, N. J., Kang, S. W. S., Lockwood, G. P., Le Couteur, D. G., Cogger, V. C. Hallmarks of aging in the liver. Computational and Structural Biotechnology Journal. 17, 1151-1161 (2019).
- Bou-Nader, M., et al. Polyploidy spectrum: a new marker in HCC classification. Gut. 69 (2), 355-364 (2019).
- Gentric, G., et al. Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease. Journal of Clinical Investigation. 125 (3), 981-992 (2015).
- Schwartz-Arad, D., Zajicek, G., Bartfeld, E. The streaming liver IV: DNA content of the hepatocyte increases with its age. Liver. 9 (2), 93-99 (1989).
- Kudryavtsev, B. N., Kudryavtseva, M. V., Sakuta, G. A., Stein, G. I. Human hepatocyte polyploidization kinetics in the course of life cycle. Virchows Archiv. B, Cell Pathology Including Molecular Pathology. 64 (6), 387-393 (1993).
- Duncan, A. W., et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 467 (7316), 707-710 (2010).
- Granja, J. M., et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nature Genetics. 53 (3), 403-411 (2021).
- Bredikhin, D., Kats, I., Stegle, O. MUON: Multimodal omics analysis framework. Genome Biology. 23 (1), 42 (2022).
- Velten, B., et al. Identifying temporal and spatial patterns of variation from multimodal data using MEFISTO. Nature Methods. 19 (2), 179-186 (2022).

