Transgene Expression in Cultured Cells Using Unpurified Recombinant Adeno-Associated Viral Vectors
Summary
Recombinant adeno-associated virus (rAAV) is widely used for clinical and preclinical gene delivery. An underappreciated use for rAAVs is the robust transduction of cultured cells without the need for purification. For researchers new to rAAV, we provide a protocol for transgene cassette cloning, crude vector production, and cell culture transduction.
Abstract
Recombinant adeno-associated viral vectors (rAAV) can achieve potent and durable transgene expression without integration in a broad range of tissue types, making them a popular choice for gene delivery in animal models and in clinical settings. In addition to therapeutic applications, rAAVs are a useful laboratory tool for delivering transgenes tailored to the researcher's experimental needs and scientific goals in cultured cells. Some examples include exogenous reporter genes, overexpression cassettes, RNA interference, and CRISPR-based tools, including those for genome-wide screens. rAAV transductions are less harmful to cells than electroporation or chemical transfection and do not require any special equipment or expensive reagents to produce. Crude lysates or conditioned media containing rAAVs can be added directly to cultured cells without further purification to transduce many cell types—an underappreciated feature of rAAVs. Here, we provide protocols for basic transgene cassette cloning and demonstrate how to produce and apply crude rAAV preparations to cultured cells. As proof of principle, we demonstrate the transduction of three cell types that have not yet been reported in rAAV applications: placental cells, myoblasts, and small intestinal organoids. We discuss appropriate uses for crude rAAV preparations, the limitations of rAAVs for gene delivery, and considerations for capsid choice. This protocol outlines a simple, low-cost, and effective method for researchers to achieve productive DNA delivery in cell culture using rAAV without the need for laborious titration and purification steps.
Introduction
Elucidating the molecular bases of cellular functions often requires the expression of transgenic DNA in cell culture. To be expressed, transgenes must penetrate through a cell's selective membrane and reach the nucleus1,2. Therefore, the ability to effectively bypass the cell's physical barriers and manipulate its central processes is a necessity for applying transgenesis to uncover new biological phenomena. One approach capitalizes on the intrinsic ability of viruses to deliver and express foreign DNA3,4.
Adeno-associated virus (AAV) is one of the smallest mammalian viruses: its 4.7 kilobase (kb), single-stranded DNA genome contains two genes, rep (for replicase) and cap (for capsid), packaged inside of a 60-mer icosahedral capsid measuring 25 nm. The rep/cap genes have multiple promoters, reading frames, and splice products which encode at least nine unique proteins required for viral replication, production, and packaging5,6. Additionally, both ends of the genome contain secondary structures called inverted terminal repeats (ITRs) that are necessary for DNA replication, genome packaging, and downstream processing during transduction7,8,9,10. The ITRs are the only DNA elements that are required for the packaging of the genome into the capsid, and therefore AAV can be cloned for transgene delivery purposes by replacing the viral rep/cap genes with a researcher's choice of regulatory elements and/or genes of interest6. The resulting recombinant AAV (rAAV), with an engineered vector genome (VG), is widely used in the clinic for human gene therapy and has amassed successes11. An underappreciated use of the vector is in the laboratory; rAAVs can efficiently achieve transgene expression in cultured cells to fulfill a researcher's experimental needs12.
The most common method for producing rAAV is by triple-plasmid transfection into HEK293 or 293T cells (Figure 1). The first plasmid, commonly called the cis plasmid, contains the desired transgene flanked by ITRs (pAAV). Depending on the application, cis plasmids with common elements, such as strong promoters or CRISPR-based tools, are available for purchase. The second is the pRep/Cap plasmid that contains the wild-type AAV rep and cap genes provided in-trans—i.e., on a separate, non-ITR containing plasmid that expresses regulatory and structural elements that then interact with the cis plasmid—and is thus called the trans plasmid. In addition to physically enclosing the VG, the capsid influences cellular tropism12,13. By providing the serotype-specific cap gene in-trans, researchers are easily able to maximize transduction efficiency by choosing an optimized capsid serotype for their given target cell. Lastly, as a Dependoparvovirus, AAV requires a helper virus to activate rep/cap expression from its viral promoters, achieved by adenoviral helper genes, provided on a third plasmid such as pAdΔF614,15. After 72 h of triple-plasmid transfection, the vector can be released from producer cells into the culture media by repeated freeze/thaw cycles. The entire plate contents are then collected, and large cellular debris is removed by centrifugation; the resulting media supernatant is a crude rAAV preparation ready for downstream transductions.
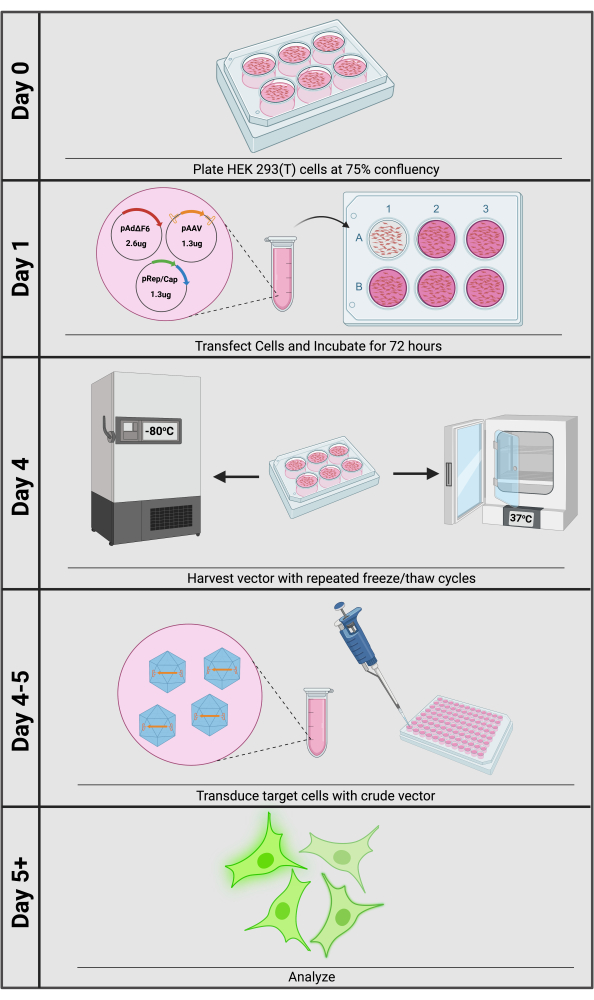
Figure 1: Overview of crude rAAV vector production. Crude rAAV production and transduction can be accomplished within 5 days. Please click here to view a larger version of this figure.
rAAV may be more favorable for transgene delivery compared to other transfection methods, which are commonly associated with cellular toxicity, low efficiency, and expensive reagents and equipment, such as for electroporation or chemical/lipid-based transfection16,17. rAAV bypasses these obstacles and often provides potent transgene expression with minimal toxicity, and minimal hands-on time. Importantly, producing rAAV and applying it in cell culture is simple and rarely requires purification of the vector from the culture media (Figure 1). Additionally, rAAV does not integrate its VG into the host genome, unlike Lentiviral transgene delivery, and thus lowers the risk of insertional mutagenesis18. Despite the potential benefits of using rAAV for transgene delivery, limitations must be considered. Importantly, the size of the transgene, including the ITRs, should not exceed 4.9 kb due to physical constraints of the capsid, thereby limiting a researcher's ability to effectively deliver large regulatory elements and transgenes. Furthermore, since rAAV is a non-integrating virus, transduction results in transient transgene expression in dividing cells and may not be practical for stable expression. However, methods using dual rAAV-delivered Cas9 and homology-directed repair (HDR) templates may be used to stably insert sequences at specific genomic loci if a researcher wishes19.
Protocol
1. Plasmid acquisition
NOTE: This protocol will clone a gene of interest (GOI) into an ITR-containing plasmid with a cytomegalovirus (CMV) promoter and a SV40 poly-adenylation sequence (pAAV.CMV.Luc.IRES.EGFP.SV40). However, this plasmid can be used without further cloning to produce rAAVs, packaging a convenient dual EGFP and Luciferase reporter. Plasmids with different regulatory elements or for different applications, such as for CRISPR-based experiments, are available online and will follow similar cloning steps as those below (see discussion for additional cloning steps). Additionally, rep/cap, or trans, plasmids for different serotypes can be used—this protocol will use rep/cap for AAV serotype 2.
- Purchase bacterial stabs containing an ITR-containing cis plasmid, an adenovirus helper plasmid, a rep/cap plasmid, and a plasmid containing a GOI. Streak the bacteria on individual agar plates containing antibiotics specific to the resistance of each plasmid. Incubate the bacteria at 30 °C overnight to grow.
NOTE: If a plasmid with a GOI is not readily available for purchase, a synthetic DNA fragment (see Materials Table) can be purchased online. Additionally, a synthetic DNA fragment can be used to skip the entire PCR process if desired. - Pick a single colony from each plate and grow in 3 mL of Luria Bertani (LB) buffer supplemented with the corresponding antibiotic at 30 °C overnight, shaking at 180 rpm. Transfer 1 mL of the bacteria into a sterile 250 mL Erlenmeyer flask containing 50 mL of LB buffer supplemented with the corresponding antibiotic. Shake at 30 °C, 180 rpm overnight.
- Transfer the bacteria to a 50 mL conical tube and centrifuge for 20 min at 3,000 x g, room temperature. Discard supernatant. Isolate the plasmid using an endotoxin-low or -free midiprep kit as per manufacturer's instructions.
NOTE: ITRs are unstable structures. To prevent ITR deletions or mutations, bacterial cells should be grown at 30 °C to slow cellular division and mitigate errors in replication. To increase plasmid yield and purity, a midiprep or maxiprep kit is ideal. It is recommended to use an endotoxin-low or -free kit to reduce downstream endotoxin contamination of cells. It is not necessary to perform plasmid isolation inside a laminar flow hood, as contamination of plasmid preparation is not likely if performed on a standard bench.
2. Cloning gene of interest into AAV ITR-containing plasmid
- Open the plasmid map containing the GOI in a DNA viewing software.
NOTE: The full cassette, including the ITRs, should not exceed 4.9 kb due to physical packaging limitations of the capsid. Additionally, PCR amplification cannot be achieved through ITRs due to secondary structures. As such, Gibson assembly is not recommended for rAAV transgene cloning.- Click the Sequence tab to reveal the full sequence of the plasmid. Scroll to the 5' most region of the GOI sequence and click on the First Nucleotide while dragging inward towards the gene body to create a forward primer. Release once a melting temperature (Tm) of ~55 °C has been reached.
- Click Forward Primer. Manually include the sequence for an EcoRI restriction site (5' GAATTC 3') at the 5' most end of the primer sequence. Additionally, add six extra random bases at 5' end of this restriction site to allow for the enzyme to efficiently interact with the DNA. Click Add Primer. Refer to Figure 2 for a representative example.
- Check the primer to ensure that it cannot form hairpins or primer dimers (except for the restriction site that is palindromic). If hairpins form, change the random sequence of the six extra bases. Additionally, ensure the primer does not bind anywhere else on the plasmid.
- Repeat the steps to create a reverse primer at the 3' most region of the GOI inward to the gene body. Click on Reverse Primer and include a NotI restriction site (5' GCGGCCGC 3').
NOTE: The GOI cannot contain EcoRI or NotI restriction sites – if so, different restriction enzymes will need to be used.
- Amplify the GOI in a 50 µL PCR reaction. Use the required individual components from Table 1.
- Place the reaction in a thermocycler and follow the cycle parameters from Table 2 for programming. If primers have different Tm, use the lower Tm for the program.
- Verify amplification of correct PCR fragment by adding 1 µL of gel loading dye (6x) to 5 µL of PCR reaction. Run a DNA ladder and the PCR mixture on a 0.8% agarose gel containing ethidium bromide and visualize with UV light.
CAUTION: Ethidium bromide is a known mutagen. - If a single, specific band appears on the gel, purify the remaining PCR product in the PCR strip tube using a column-based PCR cleanup kit as per manufacturer's instructions. If multiple bands appear (due to non-specific amplification), excise the desired band, and purify the DNA using a gel extraction kit as per manufacturer's instructions.
- Digest the purified PCR product and pAAV.CMV.Luc.IRES.EGFP.SV40 backbone plasmid in a 50 µL reaction with the required individual components from Table 3 for 1 h at 37 °C. A larger quantity of pAAV plasmid DNA is required due to expected inefficient recovery following gel extraction.
NOTE: The restriction enzymes used are high fidelity (HF) versions. Regular NotI cannot be used with CutSmart buffer. If different enzymes are being used, a different incubation temperature and buffer may be required.- Purify the digested PCR product with a column-based PCR cleanup kit as per manufacturer's instructions.
- Prepare the digested pAAV.CMV.Luc.IRES.EGFP.SV40 backbone plasmid for gel electrophoresis by adding 10 µL of gel loading dye (6x) to 50 µL of digestion reaction. Load a DNA ladder, digestion reaction, and 250 ng of undigested plasmid (negative control) on a 0.8% agarose gel containing ethidium bromide with wide-teeth wells. Visualize with UV light, excise the desired fragment (~4.5 kb), and purify the DNA using a gel extraction kit as per manufacturer's instructions.
- Ligate the digested PCR product and digested pAAV backbone plasmid in a 20 µL ligation reaction with the required individual components from Table 4. To calculate the amount of PCR products needed, use Formula 1. Typically, use a 3:1 molar ratio of PCR product (insert) to backbone (pAAV), with a mass of 50 ng backbone.
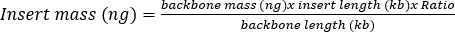 Formula 1
Formula 1 - Perform a negative control by replacing the PCR product with water. Incubate ligation reactions at 16 °C overnight, or at room temperature for 2 h.
- Thaw a vial of competent, recombination-deficient bacterial cells (such as Stbl3 cells) on ice and place 50 µL into a 1.5 mL tube. Add 3 µL of the ligation reaction directly onto the cells and incubate on ice for 30 min.
NOTE: ITRs are unstable structures and therefore require recombination-deficient bacterial strains to limit ITR deletion and recombination events. DH5α cells may be used intermittently for propagation (one or two times) but are not recommended for long-term use. ITR integrity should be checked regularly after midiprep purification.- Heat-shock the bacteria in a 42 °C water bath for 30 s then immediately transfer to ice for 2 min. Add 200 µL of LB buffer and shake for 1 h at 30 °C, 180 rpm.
- Spread 125 µL of the mixture onto an agar plate with the corresponding antibiotic and incubate overnight (~18 h) at 30 °C. Pick multiple clones and place each into a sterile plastic culture tube with 3 mL of LB containing the corresponding antibiotic. Incubate overnight shaking at 30 °C, 180 rpm.
NOTE: To prevent ITR deletions or mutations, bacterial cells should be grown at 30 °C to slow cellular division and mitigate errors in replication. Additionally, smaller colonies should be picked as those without ITRs may have a growth advantage over others. - Pipette 1.8 mL of each culture into a 2 mL tube and centrifuge at 6,000 x g for 3 min, room temperature. Isolate the DNA using a miniprep kit. Store the remaining 1.2 mL of culture at 4 °C. Verify correct clones by DNA sequencing or diagnostic restriction enzyme digest.
NOTE: Sanger sequencing of the insert is recommended, but processivity through ITRs is not achievable by standard methods. ITRs should be verified by restriction digest as described below. - Add 50 mL of LB buffer containing the corresponding antibiotic to a sterile 250 mL Erlenmeyer flask. Add 500 µL of the remaining culture to the flask and shake at 30 °C, 180 rpm until an OD600 of 0.2-0.5 is reached (~18 h). Transfer the bacteria to a 50 mL conical tube and centrifuge for 20 min at 3,000 x g, 4 °C. Isolate the DNA using an endotoxin-low or -free midiprep kit.
- Check ITR integrity with 20 µL of diagnostic restriction enzyme digest using XmaI (or SmaI). Prepare a reaction containing 500 ng plasmid, 0.5 µL of enzyme, and 1x buffer; incubate for 1 h at 37 °C. Run the reaction on a 0.8% agarose-gel. If ITRs are intact, digestion will give a band at ~2.9 kb and another band the size of the cassette. If this distinct band is not observed, the ITR may not be intact, and cloning will need to be redone.
NOTE: Plasmids containing XmaI (or SmaI) restriction sites (in addition to those in the ITRs) will have additional bands appear on the gel.
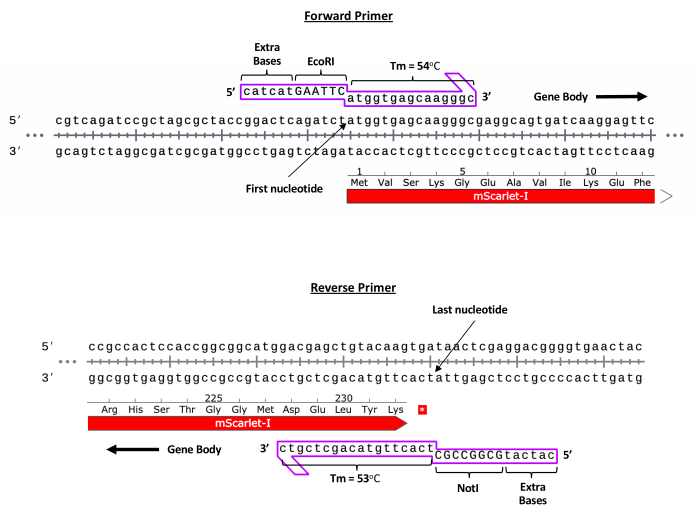
Figure 2: Representative example for primer design. Forward and reverse primer design for an mScarlet transgene (representative example). Please click here to view a larger version of this figure.
| Amount | Component |
| X µL | Template DNA (25 ng) |
| 1 µL | F primer (10 µM) |
| 1 µL | R primer (10 µM) |
| 1 µL | dNTP (10 mM) |
| 10 µL | Phusion Buffer (5x) |
| 1 µL | Phusion Polymerase |
| X µL | Water |
| 50 µL | Final Volume |
Table 1: PCR amplification reagents. The required components for a successful PCR reaction.
| Step | Temperature | Time |
| Initial Denaturation | 98 °C | 1 min |
| 30 Cycles | 98 °C | 10 s |
| Tm of Primer | 30 s | |
| 72 °C | 30 s per kb | |
| Final Extension | 72 °C | 10 min |
| Hold | 4 °C | indefinitely |
Table 2: PCR amplification programming. The required cycling parameters for a successful PCR reaction.
| Amount | Component |
| X µL | PCR product (1µg) or pAAV plasmid (3µg) |
| 5 µL | Buffer (10x) |
| 1 µL | EcoRI-HF |
| 1 µL | NotI-HF |
| X µL | Water |
| 50 µL | Final Volume |
Table 3: Digestion reagents. The required components for a successful digestion reaction.
| Amount | Component |
| X µL | Insert (X ng) |
| X µL | Backbone (50 ng) |
| 2 µL | T4 DNA Ligase Buffer (10x) |
| 1 µL | T4 DNA Ligase |
| X µL | Water |
| 20 µL | Final Volume |
Table 4: Ligation reagents. The required components for a successful ligation reaction.
3. Vector production with triple-plasmid transfection
NOTE: The following values are optimized for a single well of a 6-well plate that yields a final crude preparation volume of 2 mL. All values can be scaled up by 10x for a 15 cm plate that yields a final volume of 20 mL or scaled down by 4x for a 24-well plate with a final volume of 500 µL.
- Make a 1 µg/µL stock of polyethylenimine hydrochloride (PEI) MAX by dissolving 100 mg of PEI MAX in 100 mL of distilled water. Adjust pH to 7.1 with NaOH. Filter sterilize the mixture with a 0.22 µm filter and freeze 1 mL aliquots at -20 °C for long-term storage. Once thawed, store the reagent for 1 month at 4 °C.
- Seed 3 x 105 HEK293 or HEK293T cells into a 6-well plate with pre-warmed Dulbecco's modified eagle medium (DMEM, 4.5 g/L glucose, 110 mg/L sodium pyruvate) supplemented with 10% fetal bovine serum (FBS). Grow to ~75%-90% confluency in an incubator set at 37 °C and 5% CO2 (Figure 3).
NOTE: Cells cultured with penicillin/streptomycin (P/S) have been observed to decrease the vector production yield. Using proper sterile technique bypasses the need to use P/S, and thus it is recommended not to culture HEK293 cells with antibiotic20. If P/S is required in downstream transductions, it is recommended to add P/S to crude preparation after harvesting. - In a 2 mL tube, prepare a mixture containing 1.3 µg pAAV2/2 (Rep/Cap, serotype 2), 1.3 µg pAAV.GOI, 2.6 µg pAdΔF6, and serum-free (SF) DMEM (4.5 g/L glucose, 110 mg/L sodium pyruvate) into a total volume of 100 µL (see Supplementary Table 1 for convenient calculations).
- Prepare a negative control in a separate tube by replacing pRep/Cap with any unrelated plasmid. The negative control accounts for unpackaged pAAV.GOI plasmid present in the crude preparation that could possibly transfect cells during transduction, although rare.
NOTE: Endotoxin-low or -free maxiprep or midiprep plasmid is ideal for triple-transfection and generally produces higher titer vector compared to miniprep DNA, although miniprep DNA can be used for quick-and-dirty preliminary testing. - Add 5.2 µL of PEI MAX to the plasmid mixture; this is a plasmid:PEI mixture with a ratio of 1:1. Mix well by pulsing 10-15 times on a vortex mixer set to 7. If multiple vector preparations are produced, add PEI to each plasmid mixture at staggered time intervals of 1 min (i.e., preparation 1 at t = 0 min, preparation 2 at t = 1 min, etc.) to allow for sufficient timing to aspirate wells and dilute the reaction in the following steps.
NOTE: A 1:1 ratio of plasmid:PEI has been observed to be optimal. However, individual users can optimize this ratio for maximum efficiency. Refer to the discussion for how to perform PEI optimization (also see Supplementary Table 2). - Incubate each tube for exactly 15 min and then dilute the reaction with 1.9 mL of SF DMEM (4.5 g/L glucose, 110 mg/L sodium pyruvate) for a final volume of 2 mL. Gently pipette 2x to mix. Overmixing will disrupt Plasmid/PEI complexes and result in lower transfection efficiency.
- Aspirate media from well and add plasmid:PEI mixture gently on well sides to prevent cellular detachment. Incubate cells for 72 h at 37 °C and 5% CO2. Cell lysis and detachment during incubation is a normal process of rAAV production (Figure 4).
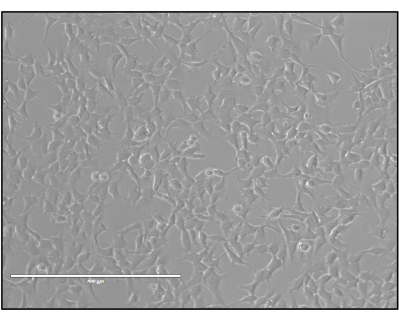
Figure 3: HEK293 cells ready for transfection. The ideal confluency (75%-90%) of HEK293 cells needed for triple-plasmid transfection. Please click here to view a larger version of this figure.
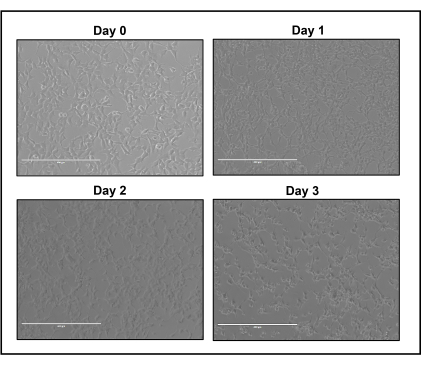
Figure 4: HEK293 cells after transfection. The appearance of HEK293 cells before and after 1, 2, or 3 days post transfection with pAAV.CMV.Luc.IRES.EGFP.SV40, pAAV2/2 , pAdΔF6, and 1:1 ratio of plasmid:PEI . Scale bars: 400 µm. Please click here to view a larger version of this figure.
4. Harvesting crude vector preparations
- Freeze the entire plate of transfected cells for 30 min at -80 °C, followed by a thaw for 30 min at 37 °C. Repeat for a total of three freeze/thaw cycles. Do not remove the lid—the plate must remain sterile inside. Plates can remain at -80 °C until ready to proceed to next steps.
NOTE: A non-humidified incubator works best for thawing, which minimizes condensation on the outside of plates that can lead to accidental contamination. - Perform the following two steps in a laminar flow hood using aseptic techniques. Mix each well by pipetting to ensure maximum disruption of cells. Transfer lysate to a 2 mL tube and centrifuge for 15 min at 15,000 x g, room temperature to remove cell debris.
- Carefully transfer supernatant to a new 2 mL tube. This crude preparation can be used immediately without titration or purification. Vector can be stored at 4 °C for several months or -20°C for years.
NOTE: If needed, titers may be calculated by quantitative PCR (qPCR) by quantifying the number of vector genomes (VG) inside of DNase-resistant particles. As such, titers are given in units of VG/mL. Vector stored for several months at 4 °C should be re-titrated before use as vector can aggregate and/or bind tube walls, reducing the titer of the preparation.
5. Transduction
- Plate the desired cell type (e.g., Huh7) in a 96-well plate at a target confluency of 50%-75%, depending on the duration of transduction. Greater confluency may result in reduced AAV transduction efficiency.
- Add vector (e.g., CMV.mScarlet) to cells without knowing the titer of the crude preparation for applications where dose is not relevant, such as testing a panel of capsids for transduction in a new cell type. Perform a 1:3 dilution series of the crude preparation in serum-free (SF) media to obtain the optimal amount of vector required for the application. Aspirate media from 96-well plate and add 50-100 µL of diluted crude preparation to wells. Incubate cells at 37 °C and 5% CO2.
NOTE: Serum may contain antibodies that can neutralize the AAV vector and reduce transduction efficiency. However, serum may be used during transduction for sensitive cell types. - Remove vector after 48 h post-transduction (hpt) and wash cells once with pre-warmed phosphate buffered saline (PBS). Vectors can also be removed and replaced with serum-containing media as soon as 2 hpt for sensitive cell types. For many applications, perform overnight incubation to transduce cells and replace wells with fresh serum-containing media in the morning. Robust cell types, such as Huh7 and U2-OS, do not require a media change.
- Terminate transduction by fixing cells in 4% paraformaldehyde (PFA) for 10 min. Various termination methods can also be used based on the application (e.g., lysis). For most cell types, peak expression occurs by 48 hpt and is thus a common experimental endpoint.
6. Variations on transduction method by cell type
NOTE: Different cell types require different culture conditions. Therefore, addition of vector for transduction should be optimized based on the needs of the cell type. Below are very specific transduction protocols for the cell types included in the representative results, illustrating some variations of transduction protocols that a researcher may wish to try for their own needs.
- Mouse small intestinal organoids
- Derive mouse small intestinal organoids (mSIOs) from a small intestinal crypt prep from C57BL/6J mice. Embed organoids in 90% basement membrane matrix and culture in 24-well plates containing either organoid growth medium (without antibiotics) or pre-transduction medium (50% Wnt3a-conditioned medium [produced in-house using L Wnt-3A cells in organoid growth medium], 10 mM nicotinamide, 10 µM ROCK inhibitor, and 2.5 µM CHIR99021) for one passage (5-7 days) before transduction at 37 °C and 5% CO2.
- Prior to transduction, rinse organoids with D-PBS, disrupt basement membrane matrix domes by pipetting, and dissociate organoids into small cell clusters by incubating them in dissociation medium for 10 min at 37 °C and further pipetting.
- Stop the cell dissociation process by adding 5% FBS in DMEM/F-12 with 15 mM HEPES, transfer cell clusters into centrifuge tubes and collect cell clusters by centrifugation for 5 min at 1000 x g, room temperature.
- Resuspend the cell clusters in transduction medium (pre-transduction medium containing 10 µg/mL polybrene or organoid growth medium containing 10 µg/mL polybrene) and transfer into 48-well plates, followed by the addition of AAV crude preparations packaging CMV.mScarlet for transduction.
- Perform transduction of organoids by centrifuging the plate for 1 h at 600 x g and 37 °C and then incubate at 37 °C for an additional 6 h. Collect transduced organoids in 1.5 mL microcentrifuge tubes, centrifuge for 5 min at 1000 x g, room temperature, and put on ice.
- After removal of the supernatant, resuspend transduced organoids in 90% basement membrane matrix in pre-transduction medium or organoid growth medium and seed droplets of 20 µL into one well of a pre-warmed chambered cover glass.
- After incubation for 10 min in the incubator, embed the droplet in 350 µL of pre-transduction medium or organoid growth medium and incubate at 37 °C in the incubator. Determine the transduction efficiency 2-5 days after transduction by imaging the transduced organoids on a confocal microscope and estimate the percentage of red fluorescent cells per organoid.
- BeWo cells
- At 24 h before transduction, plate human placental choriocarcinoma BeWo cells at 10,000 cells per well of 96-well plate. Dilute crude AAV preparations packaging CMV.mScarlet to 1:1 in serum-free F12-K BeWo medium up to a total volume of 50 µL per well. Aspirate medium from well and replace with 50 µL diluted AAV per well.
- Incubate cells at 37 °C overnight (~18 h) and then add 100 µL of F12-K BeWo medium containing 10% FBS to bring up the total volume to 150 µL. Incubate cells for an additional 6 h for a total of 24 h post-transduction (hpt).
- For imaging, stain live cells with Hoechst dye for 30 min in F12-K medium, then exchange the medium for phenol-free DMEM containing 10% FBS, 1x glutamax supplement, and 1x sodium pyruvate supplement for imaging. Conduct live-cell imaging on a confocal microscope using DAPI (for Hoechst imaging) and mCherry (for mScarlet imaging) filter sets with 37 °C and 5% CO2.
- Hepa1-6 and Huh7 cells
- Plate murine Hepa1-6 liver cells and human Huh7 liver cells in DMEM (4.5 g/L glucose, 110 mg/L sodium pyruvate, 10% FBS) at 5,000 cells per well of a 96-well plate 24 h before transduction.
- Add 50 µL of undiluted crude AAV preparations packaging CMV.mScarlet directly to cells and incubate at 37 °C for 48 h. Wash cells once in PBS, fix in 4% PFA, and stain with Hoechst dye for 10 min. Conduct imaging on a confocal microscope using the DAPI (for Hoechst imaging) and mCherry (for mScarlet imaging) filter sets.
- C2C12 and HSkMC cells
- Plate undifferentiated murine C2C12 myoblasts and undifferentiated human primary skeletal muscle cell (HSkMC) myoblasts at 5,000 cells per well of a 96-well plate, 72 h before transduction.
- Dilute crude AAV preparations packaging CMV.mScarlet to 1:1 in DMEM (4.5 g/L glucose, Penn/Strep, 20% FBS) for C2C12 myoblasts or skeletal muscle growth media for HSkMC myoblasts. Differentiate HSkMC myoblasts to myotubes by changing the media to differentiation media.
- Incubate myoblasts and myotubes in diluted AAV preparations for 24 h at 37 °C. Stain live cells with Hoechst dye for 10 min and image on a confocal microscope using the DAPI (for Hoechst imaging) and mCherry (for mScarlet imaging) filter sets.
Representative Results
Finding optimal capsid for transducing cultured cells of interest
A variety of cell types were transduced to determine the tropism of various capsid serotypes (Figure 5). The natural serotypes AAV214, AAV421, AAV522, AAV823, AAV924, and the engineered capsid variants Anc8025, DJ26, LK0327, and KP128 were packaged with a vector genome expressing mScarlet under a CMV promoter, cloned using the methods provided in this protocol. Untitrated crude vector preparations were diluted in the appropriate culture media and cells were transduced for 24+ h and imaged (Figure 5B). Transduction efficiency was calculated by the proportion of total cells that were mScarlet+, or the proportion of cells in a single z-plane that were mScarlet+ (for mouse small intestinal organoids; Figure 5C). Transduction efficiencies (mScarlet+ cells) are not provided for differentiated C2C12 and HSkMC myotubes, but rather for undifferentiated C2C12 and HSkMC myoblasts (Figure 5A).
AAV2 and KP1 were the most potent serotypes across cell lines tested (Figure 5A). It was also observed that similar cell types originating from different species are transduced at varying efficiencies. For example, Hepa1-6 (a murine-derived liver cell line) exhibits a marked decrease in transduction compared to Huh7 (a human-derived liver cell line) when using AAV2 (Figure 5B). Moreover, different media conditions play an important role during transduction. Mouse small intestinal organoids (mSIOs) cultured in pre-transduction media are less effectively transduced compared to those cultured in organoid growth media (Figure 5C). Additionally, AAV2 efficiently transduces undifferentiated HSkMC myoblasts and differentiated HSkMC myotubes (Figure 5B), though quantitative analysis of myotubes is difficult and is therefore not provided.
The high transduction efficiencies observed for various cell types in Figure 5 shows that crude vector preparations can be used effectively to transduce a variety of cell types without further steps such as purification and titration. However, careful consideration should be given when choosing a capsid to maximize transgene delivery. Many other cell lines and capsids have been previously tested and are published, though with purified vector preparations12.
A vector particle independent effect of media conditions can affect the efficiency of transduction. However, it is generally accepted that tropism (i.e., cell-type specific uptake and expression of vector) is a property that is capsid specific29. A comparison between dose-matched crude and purified preparations has not yet been observed to affect cellular tropism. Therefore, crude vector preparations can be used in a similar manner as purified vectors in cell culture.
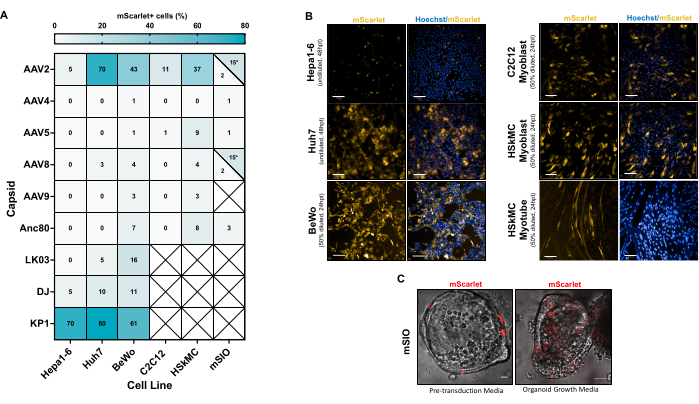
Figure 5: Crude vector transduction of various cell types. (A) Percentage of mScarlet+ following the addition of various AAV vectors containing an mScarlet transgene. (B) Cells expressing mScarlet delivered from AAV serotype 2. Scale bars: 100 µm (Hepa1-6, Huh7, C2C12, and HSkMC), 200 µm (BeWo), 20 µm (mSIO). Abbreviations: hpt = hours post transduction. (C) Mouse small intestinal organoids (mSIOs) were either rAAV treated in pre-transduction media or organoid growth media. Please click here to view a larger version of this figure.
Supplementary Table 1: Triple plasmid transfection worksheet. Please click here to download this File.
Supplementary Table 2: PEI optimization worksheet. Please click here to download this File.
Discussion
Cloning
The cloning protocol is not limited to the pAAV.CMV.Luc.IRES.EGFP.SV40 plasmid used above and can be easily altered based on a researcher's experimental needs. Many ITR-containing plasmids are readily available online for purchase. For example, plasmids containing both Cas9 and an sgRNA cloning site are available but require few additional steps such as oligonucleotide annealing and PNK treatment30. Additionally, plasmids containing a multiple cloning site (MCS) with only ITRs and no inner regulatory elements can be found31. If different plasmids are to be used, the restriction enzymes (RE) used for digestion are typically the only elements that may need to be changed in this protocol. However, a limitation of rAAV is its limited cargo capacity. Due to physical limitations of the capsid, the vector genome should not exceed 4.9 kb, including the ITRs.
When isolating plasmid from bacteria, it is critical to use an endotoxin-low or -free midiprep or maxiprep kit to mitigate harm to cells during triple-plasmid transfection or transduction. Plasmid from miniprep kits often contain higher impurities, reduced concentrations, and fewer supercoiled DNA, all of which can affect the downstream production of rAAV and is thus not recommended.
It is critical to understand the structure and properties of ITRs during cloning. First, it is extremely difficult to use PCR through the ITR. Cloning designs that require PCR amplification through ITRs should be avoided, and additionally limit the use of the Gibson assembly cloning technique. As such, restriction enzyme cloning is the preferred method for cloning into ITR-containing plasmids. Furthermore, certain primers for Sanger sequencing may not be compatible if the sequenced region contains the ITR. Instead, it is recommended to use primers that sequence away from the ITRs and into the vector genome body to get more precise sequencing results. Second, ITRs are prone to deletions, rearrangements, and mutations when transformed into bacteria for plasmid amplification32,33. To mitigate these events, it is recommended to use recombination-deficient competent bacterial strains, such as Stbl3, and to incubate them at 30 °C to slow down cellular divisions. Last, it has been observed that smaller colonies may correspond with clones without rearrangements or deletions, as those without ITRs may confer a growth advantage and be larger. Therefore, it is recommended to pick colonies that are small.
Vector production
The successful production of rAAV vector can be affected by multiple elements. One critical factor is the health of HEK293 or 293T cells used for transfection. Generally, low passage numbers are ideal, as highly passaged cells may exhibit genotypic and phenotypic variances that can reduce rAAV titers. Additionally, the density of the seeded cells should be 75%-90% confluency for effective production. Sparse cells generate low vector yields because there are less cells available to produce vectors, while overgrown cells will not be efficiently transfected.
Variations between reagent lots, cell stocks, and general lab-to-lab variability contribute to differences in transfection efficiencies and production titer. One optimizable factor that can lead to titer improvements is the plasmid:PEI ratio in transfection reactions. It is critical to use fresh (<1 month old) PEI MAX. It is recommended that a plasmid:PEI ratio of 1:1 be used as a starting point, and if transfection or transduction efficiency appears poor, test several different ratios. Titer optimization is easiest if using a transgene with a visual readout, such as the CMV.Luc.IRES.EGFP reporter trangene used herein as starting material for cloning. To perform the optimization, follow protocol step 3 using a 12-well plate and scaling down the plasmid masses and reagent volumes by two (final plasmid mass is 2.6 µg). Adjust the PEI volume accordingly to correspond to ratios ranging from 1:0.75 to 1:3, with increasing increments of 0.25 (Figure 6). Dilute each reaction with 950 µL of SF media after 15 min. For convenience, a master mix containing the triple plasmids can be made and individually pipetted into 1.5 mL tubes prior to adding PEI-see Supplementary File 2. Harvest the vector, transduce cells of interest, and image. The well with the highest transduction efficiency (proportion of GFP+ cells) corresponds to the highest titer and most optimal ratio of PEI:DNA.
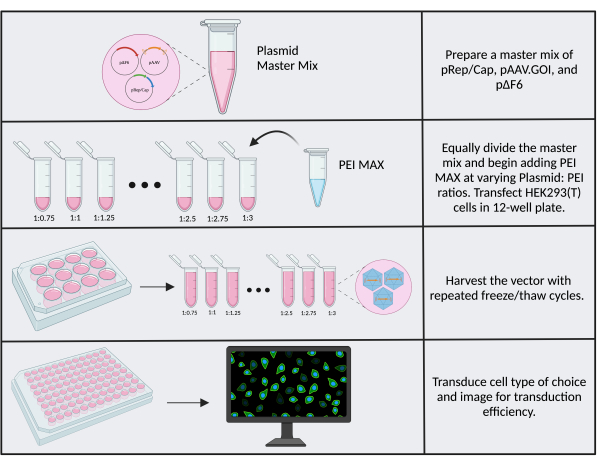
Figure 6: PEI optimization workflow. Schematic of the steps required for PEI optimization. Multiple ratios of plasmid: PEI are tested to determine the optimal ratio. Please click here to view a larger version of this figure.
Harvest and titer considerations
The freeze/thaw technique used to harvest rAAV vector effectively lyses HEK293 cells in a manner compatible with the direct use of the clarified lysate to transduce cultured cells. Certain rAAV serotypes, such as AAV1, AAV8, and AAV9 are released from cells during vector production and can be harvested from the cultured cell medium without freeze/thaw cycles34. The method described here typically yields titers on the order of 1 x 1010 VG/mL when using AAV2 capsids, and 1 x 1011 VG/mL for AAV8. While higher titers can be achieved by detergent or other chemical-based lysis, these are harmful to cells in downstream use and require rAAVs to be further purified from the lysate. Lower titer is one tradeoff a researcher should consider when determining whether crude preparations are appropriate for their research needs, however, the marginally lower titers produced by the methods described here can transduce many cell types very well (see representative results). In addition to transfection efficiency and cell health, vector titers vary depending on the capsid used during rAAV production and the size and sequence of the transgene within the VG35.
When harvesting crude vector preparations, plasmid DNA that was used during triple-plasmid transfection may be present and, although rare, result in downstream transfection during transduction. Furthermore, unpackaged VGs may bind to the exterior of capsids and invoke an innate immune response to naked and foreign single-stranded DNA36,37. Therefore, sensitive cell types may require vector preparations to be DNase digested and purified to remove unpackaged VGs and plasmid.
If one wishes to calculate the titer of a crude preparation, qPCR can be performed to quantify the number of packaged VG inside DNase-resistant particles (DRP). Briefly, a small amount of crude preparation is DNase-digested to remove plasmid DNA, contaminating nucleic acids, or partially packaged VG. The sample is then subject to qPCR and the protected VG inside of DRPs is quantified, resulting in a titer with units of vector genome per mL of crude preparation38. It is not recommended to perform vector titration using ELISA-based assays that quantify capsid titers. Compared to wild-type AAV virus, rAAV suffers from a proportion of empty and partially packaged capsids39. ELISA will quantify all capsids regardless of their genome contents and will overestimate the transducible units present in a preparation, which requires a packaged VG.
Transduction considerations
Many factors influence rAAV transductions and proper considerations should be made for any new experiment. Depending on the promoter driving transgene expression, expression onset can occur as early as 4 h post-transduction (hpt), and peak expression is typically achieved by 48 hpt. It is important to keep in mind the duration of time from the initial seeding of cells to the experimental endpoint. This is to estimate the starting confluency of the cells and ensure that they do not overgrow by the end of the experiment. If cells become overconfluent, cellular behavior may be altered due to a stress response and can confound experimental results. Some cell types, like U2-OS, can tolerate overgrowth/contact inhibition quite well. Additionally, they can withstand long periods (48 h+) in serum-free conditioned medium—the product of this production protocol. However, sensitive cell types may require serum addition or dilution of the crude preparation with special growth medium to maintain health during transduction. A slightly reduced transduction efficiency from using serum-containing media is a potential tradeoff for cell health and should be considered by the researcher.
Typically, for rapidly dividing cells, a starting confluency of around 50% is optimal for applications that will be terminated 48 hpt. However, confluency can be adjusted accordingly based on the needs of the experiment. It is not recommended to transduce monolayer-type immortalized cell lines over 75% confluency due to decreased transduction efficiencies. Most cultured cell types are successfully transduced and healthy after overnight incubation with crude rAAV preparations, followed by a change to fresh serum-containing media in the morning.
Capsid serotype is an important factor to consider when producing rAAV to transduce a target cell, as the capsid is the primary determinant of cellular tropism and subsequent transgene expression13. AAV2 is a widely used serotype due to its ability to effectively transduce many types of cultured cells12. This property of AAV2 may be attributed to heparin sulfate proteoglycans (HSPGs) serving as the primary attachment factor for AAV2 and the high levels of HSPGs on cultured cells from the adaptation to growing in a dish40. Other capsids, such as AAV9, are less effective at transducing broad cell types and may be explained by their reliance attachment factors that are not expressed in this setting41. Therefore, we recommend AAV2 as a first-choice capsid in cultured cells if a desired target cell has not been previously tested with rAAV in the literature.
Please note that a major limitation of crude vector preparations is that they are inappropriate for transducing animal models. In vivo studies require preparations to be purified and undergo quality assessment.
Transgene expression and potential integration considerations
rAAVs do not reliably result in permanent expression of the transgene. Over time, VGs can become silenced and transgenic expression may be shut down following several passages42. Additionally, the majority of VGs remain episomal, and rAAVs do not contain the viral Rep proteins that would mediate frequent integration into the host genome as in a wild-type viral lysogenic infection or promote replication of VGs43. As a result, episomes in transduced cells will eventually be diluted out among daughter cells through divisions.
Basal-level integration is a possibility for all delivered transgenic DNA material. However, ITR-containing VGs are prone to integration at a higher frequency44. Therefore, permanent expression of a transgene may be observed in a small subset of cells. Users should consider this possibility especially when using rAAV to deliver DNA-cutting enzymes, such as Cas9, as double-stranded breaks may result in an even larger frequency of integration and permanent expression45. While this makes rAAV a good candidate for delivering homology directed repair templates for endogenous tagging or gene addition, the possibility of Cas9 insertion should be considered19,46.
Declarações
The authors have nothing to disclose.
Acknowledgements
We thank Robert Tjian and Xavier Darzacq for their support and use of laboratory equipment. We thank Mark Kay for his gift of the KP1 and LK03 rep/cap plasmids, and Luk Vandenberghe for AAV4 rep/cap plasmid. Funding was provided by the Howard Hughes Medical Institute (34430, R. T.) and the California Institute for Regenerative Medicine Training Program EDUC4-12790. N.W. acknowledges funding from the Berkeley Stem Cell Center via a Siebel postdoctoral fellowship and from the German Research Foundation (DFG) via a Walter Benjamin fellowship.
Materials
| Snapgene DNA viewing sofrware | Snapgene | ||
| pAAV.CMV.Luc.IRES.EGFP.SV40 | AddGene | 105533 | |
| pAAV2/2 (Rep/Cap) | AddGene | 104963 | |
| pAdΔF6 | AddGene | 112867 | |
| LB Agar Carbenicillin | Sigma-Aldrich | L0418 | |
| Boekel Scientific Economy Digital Incubator | Boekel Scientific | 133000 | |
| LB medium, powder | MP Biomedicals | 113002042 | |
| Carbencillin (Disodium) | GoldBio | C-103-5 | |
| New Brunswick I26 Shaker | Eppendorf | M1324-0000 | |
| 50 mL Centrifuge Tubes | Corning | 430828 | |
| Centrifuge 5810 R | Eppendorf | 22627040 | |
| QIAGEN Plasmid Plus Midi Kit | Qiagen | 12945 | |
| PCR PULL-APART 8-TUBE STRIPS | USA Scientific | 1402-3900 | |
| Mastercycler nexus | Eppendorf | 6333000022 | |
| dNTP | Thermo Fisher Scientific | 18427013 | |
| 5x Phusion Buffer | NEB | B0518S | Provided with purchase of Phusion Polymerase |
| Phusion High-Fidelity DNA Polymerase | NEB | M0530S | |
| Gel Loading Dye, Orange (6X) | NEB | B7022S | |
| DNA Clean & Concentrator-100 | Zymo | D4029 | |
| Zymoclean Gel DNA Recovery Kit | Zymo | D4001 | |
| NotI-HF | NEB | R3189S | |
| EcoRI-HF | NEB | R3101S | |
| CutSmart Buffer (10x) | NEB | B6004S | Provided with purchase of restriction enzyme |
| UltraPure Agarose | Thermo Fisher Scientific | 16500100 | |
| Ethidium Bromide | Sigma-Aldrich | E1510 | |
| T4 DNA Ligase | NEB | M0202S | |
| T4 DNA Ligase Reaction Buffer | NEB | B0202S | Provided with purchase of T4 Ligase |
| Eppendorf Safe-Lock Tubes (1.5mL) | Eppendorf | 22363204 | |
| Precision Microprocessor Water Bath | Thermo Scientific | 51221046 | |
| Sterile Plastic Culture Tubes | Fisher Scientific | 149566B | |
| 2.0 mL Microcentrifuge Tube | Thomas Scientific | 1149Y01 | |
| ZR Plasmid Miniprep – Classic | Zymo | D4015 | |
| Xma1 | NEB | R0180S | |
| Sma1 | NEB | R0141S | |
| HEK 293T cells | ATCC | CRL-3216 | |
| Falcon 6-well | Corning | 353046 | |
| Gibco DMEM, high glucose, pyruvate | Thermo-Fisher | 11995065 | |
| Sanyo MCO-18AIC(UV) CO2 Incubator | Marshall Scientific | MCO-18AIC | |
| PEI MAX (Polyethylenimine Hydrochloride) | Polysciences | 24765-100 | |
| Mixer Vortex Genie 2 | Electron Microscopy Sciences | 102091-234 | |
| Sanyo Ultra Low Freezer | Sanyo | 14656-15267-16219 | |
| INCU-Line IL 10 with transparent window | VWR | 390-0384 | |
| Eppendorf Microcentrifuges | Eppendorf | 05-400-005 | |
| Falcon 96-well | Corning | 353072 | |
| C57BL/6J mice | JAX | strain #000664 | |
| organoid growth medium | STEMCELL Technologies | 6005 | |
| L Wnt-3A cells | ATCC | CRL-2647 | |
| nicotinamide | Sigma | N0636-100G | |
| ROCK inhibitor | STEMCELL Technologies | 72302 | |
| CHIR99021 | STEMCELL Technologies | 72052 | |
| Corning Matrigel Growth Factor Reduced (GFR) Basement Membrane | Fisher | 356231 | |
| 24-well plate | Fisher | 08-772-1 | |
| D-PBS | Thermo Fisher Scientific | 14-190-250 | |
| TrypLE Express | Fisher | 12604013 | |
| DMEM/F-12 with 15 mM HEPES | STEMCELL Technologies | 36254 | |
| polybrene | Millipore Sigma | TR-1003-G | |
| 48-well plates | Fisher | 08-772-3D | |
| Thermo Scientific Nunc Lab-Tek Chambered Coverglass | Fisher | 12-565-470 | |
| BeWo cells | ATCC | CCL-98 | |
| F-12K Medium | ATCC | 30-2004 | |
| Hepa1-6 | ATCC | CRL-1830 | |
| Huh7 | UC Berkeley BSD Cell Culture Facility | HUH-7 | |
| C2C12 | ATCC | CRL-1772 | |
| HSkMC | ATCC | PCS-950-010 | |
| Skeletal Muscle Cell Growth Medium | Sigma | C-23060 | |
| Skeletal Muscle Differentiation Medium | Sigma | C-23061 | |
| Invitrogen EVOS Digital Color Fluorescence Microscope | Fisher Scientific | 12-563-340 | |
| Perkin Elmer Opera Phenix | Perkin Elmer | HH14001000 | |
| PhenoPlate 96-well | Perkin Elmer | 6055302 | |
| DMEM, high glucose, no glutamine, no phenol red | Thermo-Fisher | 31053028 | |
| GlutaMAX Supplement | Thermo-Fisher | 35050079 | |
| Sodium Pyruvate | Thermo-Fisher | 11360070 | |
| pAAV2/5 (Rep/Cap) | Addgene | 104964 | |
| pAAV2/8 (Rep/Cap) | Addgene | 112864 | |
| pAAV-DJ-N589X (Rep/Cap) | Addgene | 130878 | |
| pAAV2/9n (Rep/Cap) | Addgene | 112865 | |
| pAnc80L65AAP | Addgene | 92307 | |
| KP1 (rep/cap) | gifted by Professor Mark Kay (Stanford University) | ||
| LK03 (rep/cap) | gifted by Professor Mark Kay (Stanford University) | ||
| pAAV4 (rep/cap) | gifted by Professor Luk Vandenberghe (Harvard Medical School) | ||
| pAAV.Cas9.sgRNA | Addgene | 61591 | |
| pAAV.MCS | Addgene | 46954 | |
| gBlock (synthetic DNA fragement) | IDT |
Referências
- Pillay, S., et al. Corrigendum: An essential receptor for adeno-associated virus infection. Nature. 539 (7629), 456 (2016).
- Nicolson, S. C., Samulski, R. J. Recombinant adeno-associated virus utilizes host cell nuclear import machinery to enter the nucleus. Journal of Virology. 88 (8), 4132-4144 (2014).
- Lundstrom, K. Viral Vectors in Gene Therapy. Diseases. 6 (2), 42 (2018).
- Daya, S., Berns, K. I. Gene therapy using adeno-associated virus vectors. Clinical Microbiology Reviews. 21 (4), 583-593 (2008).
- Ling, C., et al. The Adeno-Associated Virus Genome Packaging Puzzle. Journal of Molecular and Genetic Medicine. 9 (3), 175 (2015).
- Maurer, A. C., Weitzman, M. D. Adeno-Associated Virus Genome Interactions Important for Vector Production and Transduction. Human Gene Therapy. 31 (9-10), 499-511 (2020).
- Srivastava, A. Replication of the adeno-associated virus DNA termini in vitro. Intervirology. 27 (3), 138-147 (1987).
- Wang, X. S., Ponnazhagan, S., Srivastava, A. Rescue and replication of adeno-associated virus type 2 as well as vector DNA sequences from recombinant plasmids containing deletions in the viral inverted terminal repeats: selective encapsidation of viral genomes in progeny virions. Journal of Virology. 70 (3), 1668-1677 (1996).
- Earley, L. F., et al. Adeno-Associated Virus Serotype-Specific Inverted Terminal Repeat Sequence Role in Vector Transgene Expression. Human Gene Therapy. 31 (3-4), 151-162 (2020).
- Yang, J., et al. Concatamerization of adeno-associated virus circular genomes occurs through intermolecular recombination. Journal of Virology. 73 (11), 9468-9477 (1999).
- Au, H. K. E., Isalan, M., Mielcarek, M. Gene Therapy Advances: A Meta-Analysis of AAV Usage in Clinical Settings. Frontiers in Medicine. 8, 809118 (2021).
- Ellis, B. L., et al. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virology Journal. 10, 74 (2013).
- Zincarelli, C., Soltys, S., Rengo, G., Rabinowitz, J. E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Molecular Therapy. 16 (6), 1073-1080 (2008).
- Atchison, R. W., Casto, B. C., Hammon, W. M. Adenovirus-Associated Defective Virus Particles. Science. 149 (3685), 754-756 (1965).
- Meier, A. F., Fraefel, C., Seyffert, M. The Interplay between Adeno-Associated Virus and its Helper Viruses. Viruses. 12 (6), 662 (2020).
- Djurovic, S., Iversen, N., Jeansson, S., Hoover, F., Christensen, G. Comparison of nonviral transfection and adeno-associated viral transduction on cardiomyocytes. Molecular Biotechnology. 28 (1), 21-32 (2004).
- Batista Napotnik, T., Polajzer, T., Miklavcic, D. Cell death due to electroporation – A review. Bioelectrochemistry. 141, 107871 (2021).
- McCarty, D. M., Young, S. M., Samulski, R. J. Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annual Review of Genetics. 38, 819-845 (2004).
- Yang, Y., et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nature Biotechnology. 34 (3), 334-338 (2016).
- Stacey, G. N. Cell culture contamination. Methods in Molecular Biology. 731, 79-91 (2011).
- Parks, W. P., Melnick, J. L., Rongey, R., Mayor, H. D. Physical assay and growth cycle studies of a defective adeno-satellite virus. Journal of Virology. 1 (1), 171-180 (1967).
- Bantel-Schaal, U., zur Hausen, H. Characterization of the DNA of a defective human parvovirus isolated from a genital site. Virology. 134 (1), 52-63 (1984).
- Gao, G. P., et al. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proceedings of the National Academy of Science United States of America. 99 (18), 11854-11859 (2002).
- Gao, G., et al. Clades of Adeno-associated viruses are widely disseminated in human tissues. Journal of Virology. 78 (12), 6381-6388 (2004).
- Zinn, E., et al. In Silico Reconstruction of the Viral Evolutionary Lineage Yields a Potent Gene Therapy Vector. Cell Reports. 12 (6), 1056-1068 (2015).
- Grimm, D., et al. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. Journal of Virology. 82 (12), 5887-5911 (2008).
- Lisowski, L., et al. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 506 (7488), 382-386 (2014).
- Pekrun, K., et al. Using a barcoded AAV capsid library to select for clinically relevant gene therapy vectors. JCI Insight. 4 (22), e131610 (2019).
- Colon-Thillet, R., Jerome, K. R., Stone, D. Optimization of AAV vectors to target persistent viral reservoirs. Virology Journal. 18 (1), 85 (2021).
- Ran, F. A., et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 520 (7546), 186-191 (2015).
- Britton, S., Coates, J., Jackson, S. P. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. The Journal of Cell Biology. 202 (3), 579-595 (2013).
- Bi, X., Liu, L. F. DNA rearrangement mediated by inverted repeats. Proceedings of the National Academy of Science United States of America. 93 (2), 819-823 (1996).
- Samulski, R. J., Berns, K. I., Tan, M., Muzyczka, N. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proceedings of the National Academy of Science United States of America. 79 (6), 2077-2081 (1982).
- Vandenberghe, L. H., et al. Efficient serotype-dependent release of functional vector into the culture medium during adeno-associated virus manufacturing. Human Gene Therapy. 21 (10), 1251-1257 (2010).
- Sommer, J. M., et al. Quantification of adeno-associated virus particles and empty capsids by optical density measurement. Molecular Therapy. 7 (1), 122-128 (2003).
- Zhu, J., Huang, X., Yang, Y. The TLR9-MyD88 pathway is critical for adaptive immune responses to adeno-associated virus gene therapy vectors in mice. The Journal of Clinical Investigation. 119 (8), 2388-2398 (2009).
- Wagner, H., Bauer, S. All is not Toll: new pathways in DNA recognition. The Journal of Experimental Medicine. 203 (2), 265-268 (2006).
- Sanmiguel, J., Gao, G., Vandenberghe, L. H. Quantitative and Digital Droplet-Based AAV Genome Titration. Methods in Molecular Biology. 1950, 51-83 (2019).
- Grimm, D., et al. Titration of AAV-2 particles via a novel capsid ELISA: packaging of genomes can limit production of recombinant AAV-2. Gene Therapy. 6 (7), 1322-1330 (1999).
- Summerford, C., Samulski, R. J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. Journal of Virology. 72 (2), 1438-1445 (1998).
- Bell, C. L., et al. The AAV9 receptor and its modification to improve in vivo lung gene transfer in mice. The Journal of Clinical Investigation. 121 (6), 2427-2435 (2011).
- McCown, T. J., Xiao, X., Li, J., Breese, G. R., Samulski, R. J. Differential and persistent expression patterns of CNS gene transfer by an adeno-associated virus (AAV) vector. Brain Research. 713 (1-2), 99-107 (1996).
- Weitzman, M. D., Kyostio, S. R., Kotin, R. M., Owens, R. A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proceedings of the National Academy of Science United States of America. 91 (13), 5808-5812 (1994).
- Miller, D. G., Petek, L. M., Russell, D. W. Adeno-associated virus vectors integrate at chromosome breakage sites. Nature Genetics. 36 (7), 767-773 (2004).
- Hanlon, K. S., et al. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nature Communications. 10 (1), 4439 (2019).
- Porteus, M. H., Cathomen, T., Weitzman, M. D., Baltimore, D. Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Molecular and Cellular Biology. 23 (10), 3558-3565 (2003).

