Methods for Embedding Cell-Free Protein Synthesis Reactions in Macro-Scale Hydrogels
PREPARAÇÃO DO INSTRUTOR
CONCEITOS
PROTOCOLO DO ALUNO
1. Cell lysate buffer and media preparation
- Preparation of 2x YT+P agar and medium
- Prepare 2x YT+P agar by measuring out 16 g/L tryptone, 10 g/L yeast extract, 5 g/L NaCl, 40 mL/L 1 M K2HPO4, 22 mL/L 1 M KH2PO4, and 15 g/L agar. For the 2x YT+P broth, follow the previous composition but omit the agar.
- Sterilize by autoclaving the 2x YT+P.
- Preparation of the S30A buffer
- Prepare the S30A buffer with 5.88 g/L Mg-glutamate, 12.195 g/L K-glutamate, and 25 mL/L 2 M Tris, adjusted to pH 7.7 with acetic acid.
- Store the S30A buffer at 4 °C for up to 1 week.
- Preparation of the S30B buffer
- Prepare the S30B buffer with 5.88 g/L Mg-glutamate, and 12.195 g/L K-glutamate, adjusted to pH 8.2 with 2 M Tris.
- Store the S30B buffer at 4 °C for up to 1 week.
2. Cell lysate preparation (4 day protocol)
- Day 1
- Pour the 2x YT+P into agar plates supplemented with 100 µg/mL ampicillin.
- Streak E. coli from −80 °C glycerol stock, and incubate overnight at 37 °C.
- Day 2
- Inoculate a single colony from the 2x YT+P plate into 400 mL of 2x YT+P broth (supplemented with ampicillin) in a 1 L baffled flask.
- Incubate for 24 h at 37 °C with 220 rpm shaking.
- Day 3
- Measure and record the OD600 of the 24 h cultures; growth is sufficient at an OD600 of 3-4.
NOTE: Take a 1 mL aliquot of bacterial culture, and perform a serial dilution with medium (2x YT+P broth) prior to measuring the OD600 nm. Allow for the dilution effect when calculating the OD600. - Transfer the culture evenly between pre-chilled 500 mL centrifuge bottles.
- Centrifuge at 4,500 x g for 15 min at 4 °C, and then discard the supernatant.
- While centrifuging, complete the S30A buffer preparation by adding 2 mL/L of 1 M DTT to the previously prepared S30A buffer, mixing, and maintaining the buffer on ice.
- Resuspend the cell pellets in 200 mL of S30A buffer.
- Vortex the bottles vigorously until the whole pellet is completely solubilized, keeping the cells on ice as much as possible.
- Centrifuge at 4,500 x g for 15 min at 4 °C, and then discard the supernatant.
- Repeat steps 2.3.5-2.3.7 (inclusive).
- Add 40 mL of S30A buffer to each centrifuge bottle, resuspend the pellets, and transfer to pre-weighed 50 mL centrifuge tubes.
- Centrifuge at 4,000 x g for 15 min at 4 °C. Discard the supernatant by decanting, and remove the residual supernatant by pipette, keeping it on ice as much as possible.
- Re-weigh the centrifuge tubes, record the new mass, and calculate the mass of the pellets.
- Flash-freeze pellets in liquid nitrogen, and store them at −80 °C.
- Measure and record the OD600 of the 24 h cultures; growth is sufficient at an OD600 of 3-4.
- Day 4
- Thaw the pellets on ice.
- Per 1 g of pellet, add the following: 1.2 mL of S30A buffer (DTT added before use), 275 µL of protease inhibitor (8 mM stock), and 25 µL of lysozyme (10 mg/mL stock).
- Vortex vigorously until the pellets are completely solubilized with no remaining clumps, keeping them on ice as much as possible.
- In 50 mL centrifuge tubes, sonicate the cell suspensions at 120 W, 20 kHz, 30% amplitude, and 20 s/40 s on/off pulses for 5 min of sonication.
- Centrifuge at 4,000 x g for 1 h at 4 °C to pellet the cell debris.
- Transfer the supernatant to fresh 50 mL centrifuge tubes.
- Incubate tubes at 37 °C at 220 rpm for 80 min.
- Prepare dialysis materials by adding 1 mL/L of 1 M DTT to S30B buffer. Mix and add 900 mL to a 1 L sterile beaker. Add a magnetic stirrer, and keep it at 4 °C.
- Following the runoff reaction, transfer the cell extract from step 2.4.7 into 2 mL microcentrifuge tubes, and centrifuge at 20,000 x g for 10 min at 4 °C.
- Consolidate the pellet-free supernatant on ice in a 50 mL centrifuge tube.
- Determine the total volume of cell extract produced, and hydrate the necessary number of 10K MWCO dialysis cassettes by submerging them in S30B for 2 min.
- Load the cassettes via a syringe with up to 3 mL of extract. Dialyze up to three cassettes per 1 L beaker, stirring at 4 °C for 3 h.
- After the dialysis is complete, which clarifies the extract, transfer the extract to 2 mL micro-centrifuge tubes. Centrifuge at 20,000 x g for 10 min at 4 °C.
- Consolidate the clarified extract into a fresh centrifuge tube on ice, and vortex briefly to mix.
- Determine the protein concentration of the extract using a fluorometer.
- Dilute the extract to a concentration of 44.5 mg/mL using pre-chilled ddH2O.
- Aliquot into 200 µL aliquots, flash-freeze in liquid nitrogen, and store at −80 °C.
3. Preparation of lyophilized hydrogels (Method A)
- Weigh 0.75 g of agarose, add ddH2O to a volume of 100 mL, and microwave in 30 s bursts at high temperatures to create molten agarose stock.
- Using a pipette, transfer 50 µL of molten agarose to a 1.5 mL microcentrifuge tube, and allow the gel to polymerize for 20-30 min (polymerization occurs faster if the gels are transferred to a fridge).
- Flash-freeze the microcentrifuge tubes, containing the polymerized gels, in liquid nitrogen.
- Remove the lid of the centrifuge tubes, cover the centrifuge tube openings with wax film, and pierce the film several times with a needle or pipette tip.
- Transfer the microcentrifuge tubes containing the polymerized gels to −80 °C storage for 1-2 h.
- Recover the centrifuge tubes from −80 °C storage, and place in a freeze-drier, ensuring the tubes are completely dry.
- Engage the freeze-drier with the following settings: temperature: −20 °C, pressure: 0.1 mbar.
- Leave the gel to freeze-dry overnight.
- Recover the gels from the freeze-drier, and place them into −80 °C storage until required.
NOTE: Gels can be stored freeze-dried for up to 1 year.
4. Preparation of the 14x energy solution stock
- Combine all the reaction components (Table 1) in a 15 mL centrifuge tube on ice to produce a 14x energy solution.
- Aliquot the 14x energy solution into 1.5 mL microcentrifuge tubes, and store at −80 °C (smaller-volume aliquots can be used).
NOTE: The 14x energy solution can be stored for several months at −80 °C if repeated freeze-thawing of the tubes is avoided.
5. Preparation of the 4x amino acid stock
- Thaw, on ice, all 20 amino acid components of an RTS Amino Acid Sampler kit. Vortex the amino acids to ensure they are completely dissolved.
- In a 50 mL centrifuge tube, combine all 20 amino acids, and add 12 mL of sterile ddH2O. Vortex until the solution becomes clear, with only a slight haze of white coloration.
- Aliquot the 4x amino acids into 1.5 mL microcentrifuge tubes (500 µL aliquots).
- Flash-freeze the 4x amino acid solution aliquots in liquid nitrogen, and move the aliquots to −80 °C storage.
6. Cell-free buffer calibration
NOTE: The CFPS buffer used for the hydrogel reactions was calibrated for optimal DTT, Mg-glutamate, and K-glutamate concentrations following the protocol in Banks et al.34 modified from Sun et al.35. The calibration reaction compositions were selected using the design of experiments (DOE) method, with seven factor levels being selected for K-glutamate (200 mM, 400 mM, 600 mM, 1,000 mM, 1,200 mM, 1,400 mM), Mg-glutamate (0 mM, 10 mM, 20 mM, 30 mM, 40 mM, 50 mM, 60 mM), and DTT (0 mM, 5, 10 mM, 15 mM, 20 mM, 25 mM, 30 mM) stocks. JMP Pro 15 was used to generate a custom DOE design (Table 2) and conduct the analysis to determine the optimal factor concentrations.
- Recover the cell-free extract from −80 °C storage, and thaw on ice.
- Thaw the 4x amino acids, 14x energy solution, 40% PEG-8000, plasmid DNA, 3 M K-glutamate, 100 mM Mg-glutamate, and 100 mM DTT stocks on ice.
- Create a calibration master mix by combining 12.5 µL of the 4x amino acids, 3.57 µL of the 14x energy solution, 2.5 µL of 40% PEG-8000, 3 µg of plasmid DNA, and 10 µL of cell-free extract (44.5 mg/mL), and make up to 35 µL per reaction with ddH2O.
- Distribute the 35 µL of master mix into the wells of a black 384-well microtiter plate.
- Following the DOE design, add the appropriate volume of Mg-glutamate, K-glutamate, and DTT to each reaction, along with ddH2O to make each reaction up to 50 µL.
- Transfer the microtiter plate to a plate reader for fluorescence detection and analysis using the plate reader settings described (for mCherry): excitation: 587 nm, emission: 610 nm, wavelength bandwidth: 12 nm, optics: top, temperature: 37 °C, with fluorescence readings being collected every 5 min for 4 h of total read time. Incubate the plates with shaking on a pulsed setting at 60 rpm.
- Upload the results into the JMP DOE design, and generate a prediction profile to determine the optimal concentration levels for K-glutamate, Mg-glutamate, and DTT based on desired response variables; in this case, the maximum fluorescence and reaction rate were the response variables.
- Combine the reagents as shown in Table 3 to create the 2X CFPS buffer
- Aliquot and store at −80 °C
7. CFPS in rehydrated lyophilized hydrogels (Method A)
NOTE: The plasmids used in the CFPS reactions were created using components from the EcoFlex modular toolkit for E. coli36 and described in Whitfield et al.11 The mCherry and eGFP were under the control of the constitutive JM23100 promoter. These components are available from Addgene.
- Recover the cell-free extract from −80 °C storage, and thaw on ice for approximately 20 min.
- Collect the 2x CFPS buffer and plasmid DNA, and thaw on ice.
- Collect the freeze-dried hydrogels, and allow them to reach room temperature for 15 min by leaving the gels to stand on the bench.
- Transfer the gels to 1.5 mL microcentrifuge tubes, trimming the gels with a scalpel if necessary to make them fit in the wells.
- In a 1.5 mL microcentrifuge tube, combine 10 µL of cell-free extract with 25 µL of 2x CFPS buffer and 4 µg of plasmid DNA; make up to a total volume of 50 µL with ddH2O to create the CFPS solution.
- Pipette the CFPS solution onto the freeze-dried hydrogels.
- Allow the gels to soak in the CFPS system for 5-10 min at room temperature.
- Transfer the gels to a black 384-well microtiter plate using a spatula.
- Transfer the microtiter plate to a plate reader for fluorescence detection and analysis using the plate reader settings described in step 6.6. Representative results are available in previous studies31,33.
8. Cell-free protein synthesis in deployable hydrogels (Method B)
- Recover cell-free extract from −80 °C storage, and thaw on ice for approximately 20 min.
- Collect the plasmid DNA, and thaw on ice.
- In a 1.5 mL microcentrifuge tube on ice, combine 10 µL of cell-free extract with 3 µg of plasmid DNA and 25 µL of 2x CFPS buffer, making up to a total volume of 37.5 µL.
- Measure 3 g of agarose, and add to 100 mL of ddH2O buffer to create 3% agarose.
- Microwave the 3% agarose in 30 s bursts at high power.
- Pipette 12.5 µL of molten agarose into 1.5 mL microcentrifuge tubes containing CFPS mix to a total volume of 50 µL.
- Allow the molten agarose to cool, but not polymerize, leaving the agarose in a heat block set to 50 °C.
- Combine the molten agarose with the CFPS solution; mix via pipetting and stirring with the tip, and ensure to move quickly to avoid gel polymerization before the CFPS solution can be mixed in.
- Allow the gels to cool at room temperature and polymerize for approximately 2 min.
- Transfer the polymerized agarose to 1.5 mL microcentrifuge tubes with a spatula, and flash-freeze in liquid nitrogen.
- Place the flash-frozen hydrogels into −80 °C storage for 1 h.
- Recover the gels from storage; remove the microcentrifuge tube lids, cover the tubes with wax film, and pierce the film to allow the moisture to be dried off.
- Engage the freeze-drier with the following settings: temperature: −20 °C, pressure: 0.1 mbar, freeze drying for 18 h (overnight).
- Store the freeze-dried CFPS devices in −80 °C storage until use.
- The freeze-dried CFPS devices, with an approximate wet volume of 50 µL, can be rehydrated with 50 µL of ddH2O without excess liquid. Rehydration takes approximately 30 min.
- Transfer the gels to a black 384-well microtiter plate using a spatula.
- Transfer the microtiter plate to a plate reader for fluorescence detection and analysis using the plate reader settings described in step 6.6. Representative results are available in previous publications31,33.
Methods for Embedding Cell-Free Protein Synthesis Reactions in Macro-Scale Hydrogels
Learning Objectives
This protocol details two methods for embedding CFPS reactions into hydrogel matrices, with Figure 1 presenting a schematic overview of the two approaches. Both methods are amenable to freeze-drying and long-term storage. Method A is the most utilized methodology for two reasons. First, it has been shown to be the most applicable method for working with a range of hydrogel materials11. Second, Method A allows for the parallel testing of genetic constructs. Method B is more appropriate for the fabrication of an optimized system and field deployment. Both protocols allow many samples to be prepared in one go to aid in experimental reproducibility. This feature is also useful for the long-term development of the technology, as freeze-dried devices may be shipped in a dry state and reconstituted on site when needed.
The approach outlined in the protocol and Figure 1 can be used for the expression of single gene constructs or for the co-expression of multiple genes. The data presented in Figure 2 show the expression of both eGFP and mCherry in a 0.75% agarose gel. Confocal microscopy was used to confirm that protein expression was homogenous throughout the hydrogel, including within the internal planes. Specifically, protein synthesis was not confined to the outer edges of the hydrogel, and internal fluorescence was not the result of protein diffusion. To confirm this, by placing an eGFP-expressing hydrogel in physical contact with an mCherry-expressing hydrogel, it was possible to see protein diffusion from one hydrogel to another. The rate of diffusion between the two was insufficient to explain the extensive localization of either red or green fluorescence inside the material. This experiment also illustrates a key advantage of deploying cell-free devices in hydrogels-the device functionality can be spatially organized in a manner that is not possible in liquid cell-free reactions. In addition, for the creation of gene networks, the simultaneous synthesis of more than a single gene product is needed. The results shown in Figure 2 (bottom row) confirmed the co-expression of both mCherry and eGFP in agarose. In this work, both proteins were expressed, and there was no spatial competition between the proteins. Again, an overlay of the red and green wavelength range demonstrates the even spatial distribution of both proteins within the hydrogel.
Table 1: The 14x energy solution stocks. Please click here to download this Table.
Table 2: Design of an experimental array for the optimization of DTT, Mg-glutamate, and K-glutamate within the cell-free protein synthesis reactions. Please click here to download this Table.
Table 3: The 2x CFPS buffer components. Please click here to download this Table.
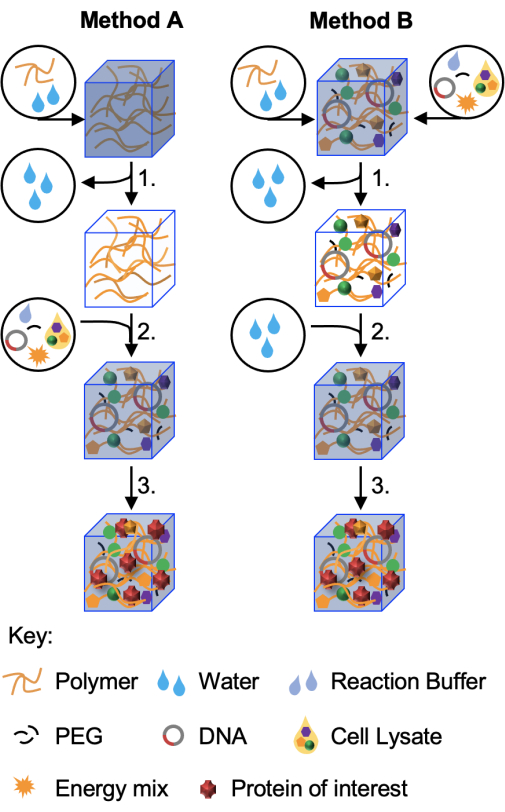
Figure 1: Schematic of the two protocols. In the first method (Method A, demonstrated in this paper) hydrogel materials are prepared first and then freeze-dried (step 1) without cell-free components. These dried hydrogels can be stored and reconstituted when required (step 2) with the correct volume of cell-free reaction prior to incubation for protein production (step 3) The variant method, Method B, incorporates all, or some, of the cell-free reaction components in the initial hydrogel fabrication. Following freeze-drying (step 1), the hydrogels may then be reconstituted in water alone or in buffer containing an analyte of interest (step 2). Protein production (step 3) continues as before. A third method, in which freeze-dried cell-free components are reconstituted with hydrogel polymers, is described in Whitfield et al.11 but has found use with only a limited number of hydrogels to date. Please click here to view a larger version of this figure.
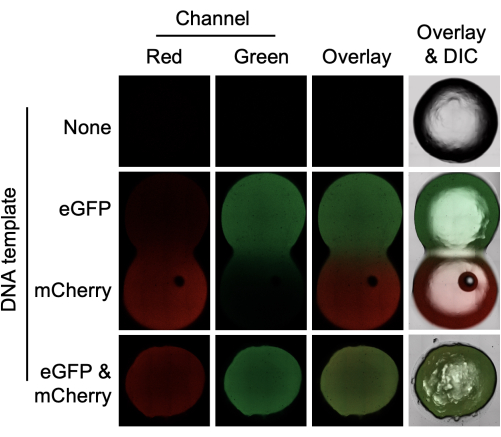
Figure 2: Cell-free protein synthesis of eGFP and mCherry in a hydrogel using E. coli cell lysates. Agarose gels (0.75%) were prepared without DNA template (top) with 4 µg of either eGFP or mCherry template (middle) or with 4 µg of both eGFP and mCherry template (bottom). The hydrogels were incubated for 4 h before confocal microscopy in the red and green channels. An overlay of the two channels is also shown, and the overlay includes the differential interference contrast (DIC) image. Hydrogels containing either eGFP or mCherry template were prepared separately but incubated in physical contact with each other. The gel diameter is 6 mm. Please click here to view a larger version of this figure.
List of Materials
| Material | |||
| 3-PGA | Santa Cruz Biotechnology | sc-214793B | |
| Acetic Acid | Sigma-Aldrich | A6283 | |
| Agar | Thermo Fisher Scientific | A10752.22 | |
| Agarose | Severn Biotech | 30-15-50 | |
| Amino Acid Sampler Kit | VWR | BTRABR1401801 | |
| ATP | Sigma-Aldrich | A8937-1G | |
| cAMP | Sigma-Aldrich | A9501-1G | |
| Coenzyme A (CoA) | Sigma-Aldrich | C4282-100MG | |
| CTP | Alfa Aesar | J14121.MC | |
| DTT | Thermo Fisher Scientific | R0862 | |
| Folinic Acid | Sigma-Aldrich | F7878-100MG | |
| GTP | Carbosynth | NG01208 | |
| HEPES | Sigma-Aldrich | H4034-25G | |
| K-glutamate | Sigma-Aldrich | G1149-100G | |
| Lysozyme | Sigma-Aldrich | L6876-1G | |
| Mg-glutamate | Sigma-Aldrich | 49605-250G | |
| NAD | Sigma-Aldrich | N6522-250MG | |
| PEG-8000 | Promega | V3011 | |
| Potassium Hydroxide (KOH) | Sigma-Aldrich | 757551-5G | |
| Potassium Phosphate Dibasic (K2HPO4) | Sigma-Aldrich | P3786-500G | |
| Potassium Phosphate Monobasic (KH2PO4) | Sigma-Aldrich | RDD037-500G | |
| Protease Inhibitor cocktail | Sigma-Aldrich | P2714-1BTL | |
| Qubit Protein concentration kit | Thermo Fisher Scientific | A50668 | |
| Rossetta 2 DE 3 E.coli | Sigma-Aldrich | 71397-3 | |
| Sodium Chloride (NaCl) | Sigma-Aldrich | S9888-500G | |
| Spermidine | Sigma-Aldrich | 85558-1G | |
| Tryptone | Thermo Fisher Scientific | 211705 | |
| Tris | Sigma-Aldrich | GE17-1321-01 | |
| tRNA | Sigma-Aldrich | 10109541001 | |
| UTP | Alfa Aesar | J23160.MC | |
| Yeast Extract | Sigma-Aldrich | Y1625-1KG | |
| Equipment | |||
| 1.5 mL microcentrifuge tubes | Sigma-Aldrich | HS4323-500EA | |
| 10K MWCO dialysis cassettes | Thermo Fisher Scientific | 66381 | |
| 15 mL centrifuge tube | Sarstedt | 62.554.502 | |
| 50 mL centrifuge bottles | Sarstedt | 62.547.254 | |
| 500 mL centrifuge bottles | Thermo Fisher Scientific | 3120-9500 | |
| Alpha 1-2 LD Plus freeze-dryer | Christ | part no. 101521, 101522, 101527 | |
| Benchtop Centrifuge | Thermo Fisher Scientific | H-X3R | |
| Black 384 well microtitre plates | Fischer Scientific | 66 | |
| Cuvettes | Thermo Fisher Scientific | 222S | |
| Elga Purelab Chorus | Elga | ##### | |
| Eppendorf Microcentrifuge 5425R | Eppendorf | EP00532 | |
| High Speed Centrifuge | Beckman Coulter | B34183 | |
| JMP license | SAS Institute | 15 | |
| Magnetic Stirrer | Fischer Scientific | 15353518 | |
| Parafilm | Amcor | PM-966 | |
| Photospectrometer (Biophotometer) | Eppendorf | 16713 | |
| Pipettes and tips | Gilson | ##### | |
| Precision Balance | Sartorius | 16384738 | |
| Qubit 2.0 Fluorometer | Thermo Fisher Scientific | Q32866 | |
| Shaking Incubator | Thermo Fisher Scientific | SHKE8000 | |
| Sonic Dismembrator (Sonicator) | Thermo Fisher Scientific | 12893543 | |
| Static Incubator | Sanyo | MIR-162 | |
| Syringe and needles | Thermo Fisher Scientific | 66490 | |
| Thermo max Q8000 (Shaking Incubator) | Thermo Fisher Scientific | SHKE8000 | |
| Varioskan Lux platereader | Thermo Fisher Scientific | VLBL00GD1 | |
| Vortex Genie 2 | Cole-parmer | OU-04724-05 | |
| VWR PHenomenal pH 1100 L, ph/mv/°c meter | VWR | 662-1657 |
Preparação do Laboratório
Synthetic gene networks provide a platform for scientists and engineers to design and build novel systems with functionality encoded at a genetic level. While the dominant paradigm for the deployment of gene networks is within a cellular chassis, synthetic gene networks may also be deployed in cell-free environments. Promising applications of cell-free gene networks include biosensors, as these devices have been demonstrated against biotic (Ebola, Zika, and SARS-CoV-2 viruses) and abiotic (heavy metals, sulfides, pesticides, and other organic contaminants) targets. Cell-free systems are typically deployed in liquid form within a reaction vessel. Being able to embed such reactions in a physical matrix, however, may facilitate their broader application in a wider set of environments. To this end, methods for embedding cell-free protein synthesis (CFPS) reactions in a variety of hydrogel matrices have been developed. One of the key properties of hydrogels conducive to this work is the high-water reconstitution capacity of hydrogel materials. Additionally, hydrogels possess physical and chemical characteristics that are functionally beneficial. Hydrogels can be freeze-dried for storage and rehydrated for use later. Two step-by-step protocols for the inclusion and assay of CFPS reactions in hydrogels are presented. First, a CFPS system can be incorporated into a hydrogel via rehydration with a cell lysate. The system within the hydrogel can then be induced or expressed constitutively for complete protein expression through the hydrogel. Second, cell lysate can be introduced to a hydrogel at the point of polymerization, and the entire system can be freeze-dried and rehydrated at a later point with an aqueous solution containing the inducer for the expression system encoded within the hydrogel. These methods have the potential to allow for cell-free gene networks that confer sensory capabilities to hydrogel materials, with the potential for deployment beyond the laboratory.
Synthetic gene networks provide a platform for scientists and engineers to design and build novel systems with functionality encoded at a genetic level. While the dominant paradigm for the deployment of gene networks is within a cellular chassis, synthetic gene networks may also be deployed in cell-free environments. Promising applications of cell-free gene networks include biosensors, as these devices have been demonstrated against biotic (Ebola, Zika, and SARS-CoV-2 viruses) and abiotic (heavy metals, sulfides, pesticides, and other organic contaminants) targets. Cell-free systems are typically deployed in liquid form within a reaction vessel. Being able to embed such reactions in a physical matrix, however, may facilitate their broader application in a wider set of environments. To this end, methods for embedding cell-free protein synthesis (CFPS) reactions in a variety of hydrogel matrices have been developed. One of the key properties of hydrogels conducive to this work is the high-water reconstitution capacity of hydrogel materials. Additionally, hydrogels possess physical and chemical characteristics that are functionally beneficial. Hydrogels can be freeze-dried for storage and rehydrated for use later. Two step-by-step protocols for the inclusion and assay of CFPS reactions in hydrogels are presented. First, a CFPS system can be incorporated into a hydrogel via rehydration with a cell lysate. The system within the hydrogel can then be induced or expressed constitutively for complete protein expression through the hydrogel. Second, cell lysate can be introduced to a hydrogel at the point of polymerization, and the entire system can be freeze-dried and rehydrated at a later point with an aqueous solution containing the inducer for the expression system encoded within the hydrogel. These methods have the potential to allow for cell-free gene networks that confer sensory capabilities to hydrogel materials, with the potential for deployment beyond the laboratory.
Procedimento
Synthetic gene networks provide a platform for scientists and engineers to design and build novel systems with functionality encoded at a genetic level. While the dominant paradigm for the deployment of gene networks is within a cellular chassis, synthetic gene networks may also be deployed in cell-free environments. Promising applications of cell-free gene networks include biosensors, as these devices have been demonstrated against biotic (Ebola, Zika, and SARS-CoV-2 viruses) and abiotic (heavy metals, sulfides, pesticides, and other organic contaminants) targets. Cell-free systems are typically deployed in liquid form within a reaction vessel. Being able to embed such reactions in a physical matrix, however, may facilitate their broader application in a wider set of environments. To this end, methods for embedding cell-free protein synthesis (CFPS) reactions in a variety of hydrogel matrices have been developed. One of the key properties of hydrogels conducive to this work is the high-water reconstitution capacity of hydrogel materials. Additionally, hydrogels possess physical and chemical characteristics that are functionally beneficial. Hydrogels can be freeze-dried for storage and rehydrated for use later. Two step-by-step protocols for the inclusion and assay of CFPS reactions in hydrogels are presented. First, a CFPS system can be incorporated into a hydrogel via rehydration with a cell lysate. The system within the hydrogel can then be induced or expressed constitutively for complete protein expression through the hydrogel. Second, cell lysate can be introduced to a hydrogel at the point of polymerization, and the entire system can be freeze-dried and rehydrated at a later point with an aqueous solution containing the inducer for the expression system encoded within the hydrogel. These methods have the potential to allow for cell-free gene networks that confer sensory capabilities to hydrogel materials, with the potential for deployment beyond the laboratory.
