Crystallization and In Situ Room Temperature Data Collection Using the Crystallization Facility at Harwell and Beamline VMXi, Diamond Light Source
Summary
We present a protocol for the crystallization of proteins using the crystallization facility in the Research Complex at Harwell and subsequent in situ X-ray crystallographic data collection from crystals within the plates at Diamond's Versatile Macromolecular Crystallography in situ (VMXi) beamline. We describe sample requirements, crystallization protocols, and data collection guidelines.
Abstract
Protocols for robotic protein crystallization using the Crystallization Facility at Harwell and in situ room temperature data collection from crystallization plates at Diamond Light Source beamline VMXi are described. This approach enables high-quality room-temperature crystal structures to be determined from multiple crystals in a straightforward manner and provides very rapid feedback on the results of crystallization trials as well as enabling serial crystallography. The value of room temperature structures in understanding protein structure, ligand binding, and dynamics is becoming increasingly recognized in the structural biology community. This pipeline is accessible to users from all over the world with several available modes of access. Crystallization experiments that are set up can be imaged and viewed remotely with crystals identified automatically using a machine learning tool. Data are measured in a queue-based system with up to 60° rotation datasets from user-selected crystals in a plate. Data from all the crystals within a particular well or sample group are automatically merged using xia2.multiplex with the outputs straightforwardly accessed via a web browser interface.
Introduction
X-ray crystallography remains a key tool for understanding protein structure and function, providing high-resolution structures of proteins or their complexes with, for example, substrates or drug candidates. In many cases, however, obtaining crystals with desirable properties – highly diffracting, crystal form amenable to soaking and without crystal pathologies such as twinning – remains a considerable bottleneck1. As suitable chemical conditions to produce protein crystals cannot, in general, be predicted, crystallization screening exploring thousands of potential chemical mixtures is standard, often aided by automation/robotics in setting screens and crystal hotels for monitoring, often remotely, the crystallization drop images that are recorded.
When crystals appear, typically they must be harvested from the crystallization environment using a nylon or Kapton loop and then, transferred to a droplet containing a cryoprotection agent (the search for which is an additional variable) before plunge-freezing into liquid nitrogen. These additional steps between crystallization and X-ray data collection can involve dehydration of the crystallization drop when its sealed environment is broken, mechanical stresses on the crystal when it is handled, and damage from the cryoprotection agents to the crystal lattice (typically resulting in increased mosaic spread) amongst other factors2. In addition, crystal harvesting is time- and labor-intensive and can lead to inhomogeneity between samples, especially when skin forms on drops during the harvesting process. The VMXi beamline gives access to useable data from crystals that are stuck to the plate, which would otherwise be discarded for data collection.
The vast majority of X-ray crystal structures are determined at 100K using the above approach, enabling straightforward crystal transport and handling and increasing crystal lifetime in the X-ray beam by orders of magnitude. There is increasing interest, however, in determining structures under non-cryogenic conditions, that is, much closer to the physiological conditions relevant to protein function2,3,4. This enables a much better appreciation of the dynamic structure of proteins, avoids amino acid conformations or loops being frozen out in functionally non-relevant states5, and enables ligand binding to be explored under conditions much closer to those in the protein's natural environment within the cell and organism6.
An alternative approach, implemented at the Versatile Macromolecular Crystallography in situ (VMXi) beamline at the Diamond Light Source synchrotron, UK, is to measure the diffraction data directly from crystals within the environment in which they have grown (i.e., within the crystallization plate), under ambient conditions and without disturbance7,8. This enables very rapid feedback from crystallization screens and optimizations to guide a user to an optimal crystal form for their requirements. It also enables high-quality room-temperature structures to be produced in an automated manner9.
This protocol assumes that a user has a highly pure protein sample ready for crystallization. We describe the user experience accessing the Crystallization Facility at Harwell to produce protein crystals and then use beamline VMXi for data collection (Figure 1).
The Crystallization Facility at Harwell
The Crystallization Facility at Harwell (CF) is located in the Research Complex at Harwell (RCaH) adjacent to the Diamond Light Source. The facility offers users a high-throughput automated laboratory for macromolecular crystallization, using robotics for crystallization screening, crystal optimization, crystal imaging, and characterization. Through close integration with the highly automated VMXi beamline, the pace of determining room temperature structures has greatly accelerated and enables the characterization of novel protein structures, protein-ligand and DNA-ligand complexes, as well as automated fragment screening (Figure 1), all under non-cryogenic conditions.
The CF pipeline is a suite of instrumentation encompassing nanoliter crystallization robots9 for the crystallization of soluble and membrane proteins, liquid handling robots to prepare commercial crystallization screens and complex custom optimization screens, and four imaging instruments (one at 4 °C and three at 20 °C for the imaging of crystallization plates (see the Table of Materials). One imager is capable of imaging lipid cubic phase (LCP) glass plates and one imager is equipped with multi-fluorescence optics (both at 20 °C).
The facility is now used widely by a broad spectrum of academic and industrial users, including the Membrane Protein Laboratory (MPL;https://www.diamond.ac.uk/Instruments/Mx/MPL.html), the XChem fragment screening facility 10, MX beamlines, the XFEL-hub, as well as the Rosalind Franklin Institute (RFI). This well-established and optimized pipeline has enabled crystallization experiments to be performed across a wide spectrum of structural biology projects. This paper describes the pipeline for crystals intended for data collection at VMXi, although crystals may also be harvested and cryo-cooled or directed to the XChem pipeline.
User access is allocated through the Diamond MX proposal system (https://www.diamond.ac.uk/Instruments/Mx/Synchrotron-Access.html) and industrial users are supported through the Diamond Industry Liaison group. All users can come to the site with their sample(s) or plates, which can be transported by hand. It is not recommended to send plates by courier as our experience suggests that drops can move away from the location in which they were dispensed, or drops may be damaged by the crystallization reservoir. Alternatively, by arrangement, users can send their protein samples to the CF, where members of staff set up crystallization experiments on their behalf. The experiments can be monitored remotely by the user by either logging on to Rock Maker Web in the case of CF or via ISPyB in the case of VMXi. Access to the CF can be carried out in an iterative fashion based on the X-ray diffraction results collected at Diamond.
Beamline VMXi at Diamond Light Source
Beamline VMXi (hereafter referred to as "the beamline") is a unique and recently developed instrument fully dedicated to room-temperature, highly automated X-ray crystallography with a focus on measuring data from crystals within suitable crystallization plates. The beamline offers a micro focus (10 x 10 µm), pink beam (band pass of <5 × 10-2ΔE/E) with a high flux of ~2 × 1013 photons/s (at 16 KeV)7. This high-flux beam, coupled with a fast detector, enables very high throughput of samples and collection of data from samples upwards of 10 µm in size.
Crystallization plates enter the beamline by being stored in a sample storage system and imaged based on the schedule provided by the user while registering the plates using the ISPyB11 interface SynchWeb12. Typically, users are advised to select a Fibonacci sequence of time points for imaging (0, 12, 24, 36, 60…7,320 h from the plate being entered into the system). The user is informed by email once a plate has been imaged. Both visible light and UV light imaging are available to users on demand. The images taken by the sample storage system are analyzed by a machine learning algorithm; this automatically locates and defines points of interest of objects that resemble crystals and registers the points of interest ready for the user to add to a queue for data collection. Users may also manually click on the visible light images to register points of interest or can click and drag a region to be analyzed by raster scan. These points are available to users to add to the queue alongside the automatically located points.
Once all samples have appropriate parameters for data collection, the plate enters a queue. When the plate reaches the top of the queue, it is automatically dispensed to the beamline. The crystallization plates are loaded from the crystal hotels into the beamline automatically by a robotic arm, and following image matching, crystallographic data sets of up to 60° rotation are measured from each selected crystal as per user defined instructions. All drops within a plate may be used for these experiments on the beamline. Data are merged from multiple crystals to produce isomorphous, optimally merged data sets in an automated manner7,9. Once all of the queued data sets are collected, the user is sent an email with a link to follow to view the data sets in ISPyB11, as in other Diamond MX beamlines. Users are also directed to the beamline web page (https://www.diamond.ac.uk/Instruments/Mx/VMXi.html).
Protocol
1. Producing crystals within in situ plates using the Crystallization Facility at Harwell
NOTE: Access to the CF is supported by a number of different routes and depends on the application of the project and user type (academic or industry). XChem and MPL projects have their own proposal application system via the User Administration System (UAS) and can be submitted either through the standard access route (including iNEXT Discovery and EUbOPEN) or BAG Access. The protocol below is specific for VMXi users.
- Proposal submission and preparation for the visit
- Provide information about the project to a BAG proposal application or add it to an active BAG proposal. There is usually a BAG coordinator, who organizes the paperwork. Alternatively, submit a rapid access proposal for access to the beamline.
- Ensure that the sample has been registered and safety validated in UAS on a proposal prior to arrival on site either by courier or in person.
- Ensure the user is registered (with FedID and password).
- Ensure the user has been added to an MX proposal as an associate in UAS by the BAG coordinator.
- Complete the beamline crystallization sample details form and send the form to VMXi@diamond.ac.uk.
- Communicate with the beamline staff about the experiment requirements and beamline availability.
- If protein samples are shipped, only send samples by prior arrangement. Refer to section 1.2 for details.
- If the user is to come onto the site to set up crystallization plates in the CF, check with the facility staff about the availability of a time slot to use the facility instrumentation and follow section 1.2.1.
- If the user brings plates onto the site, ensure that the sample is dispensed into the correct plate type and place the crystallization drops in the correct location and the correct quantity. Follow section 1.2.2. The beamline accepts only specific in situ crystallization plates (Greiner CrystalQuickX and MiTeGen In Situ-1); ensure that drops are no larger than 200 nL.
- Crystallization experiment conducted in CF
NOTE: The facility offers a number of high-throughput macromolecular crystallization methods such as vapor diffusion as well as batch crystallization under oil and LCP. It is recommended to start with 70-100 µL of pure protein and perform vapor diffusion experiments for soluble proteins with three screens using a ratio of 100 nL of protein solution and 100 nL of crystallization reservoir solution and incubating the plates at 20 °C. A number of commercial screens are available within the facility. Humidity and temperature control are available with 4 °C and 20 °C most used. Users who visit the CF receive standardized training and support operating the crystallization instrumentation and will use the settings described herein.- Shipping samples for set up at the CF
NOTE: Prior to arrival on site, the protein sample must have been validated on a proposal within the UAS system. Once the protein sample has arrived on site, members of staff will set up crystallization experiments as instructed in previous communications with the user. Confirmation will be sent via E-mail with barcode information for the experimental crystallization plates. The user will be asked to add the crystallization plates as containers to the relevant proposal. Once this has been done, the plates can be stored in automated imagers in the crystallization facility or at the beamline. ISPyB will be the interface used for interaction at the beamline.- Provide the protein sample solution at the concentration for crystallization in multiples of 25 µL aliquots. Clearly label the sample tubes containing the protein sample.
- If required, provide a protein buffer solution, ligand solution, or reservoir solution.
- Inform facility staff which screens and drop ratios should be used.
- Crystallization plate settings
NOTE: We require the crystallization drops in the Greiner CrystalQuickX and MiTeGen In Situ-1 plate to be in a particular location; plates set up elsewhere should use the following Mosquito13 settings described here.- To adjust the plate definition for MiTeGen In Situ-1, open the Mosquito SPT software and click the standard MiTeGen In Situ-1 plate definition page by clicking Setup | 96 Well | MiTeGen In Situ-1 (96 x 2 drops) (Figure 2A). Click the edit button and amend the values for sub well 2 location: X Offset to – 1.2 and Y Offset to 1.8 and for sub well 3 location: X Offset to 1.3 and Y Offset to 1.8 (Figure 2B,C).
- To adjust the plate definition for Greiner CrystalQuickX, open the Mosquito SPT software and click the standard Greiner CrystalQuickX plate definition page by clicking Setup | 96 Well | Greiner CrystalQuickX (Figure 2D). Click the edit button and amend the values for sub well 1 location: X Offset to – 1.95 and Y Offset to 1.45 and for sub well 2 location: X Offset to 1.95 and Y Offset to 1.45 (Figure 2E,F).
- Shipping samples for set up at the CF
2. Using the beamline at Diamond Light Source
NOTE: All interaction with the beamline by users is conducted remotely using the ISPyB11 interface. No physical presence at the beamline is required and data are collected using a queue-based system rather than being scheduled at a particular time. Users will have a proposal associated with their Diamond Light Source access. At the beamline, each crystallization plate is allocated a unique visit and is defined as a 'container' within ISPyB11 analogous to a puck containing samples at 100 K. Optimization screens cannot be created using the SynchWeb interface and as such, information is typically added into the comments section (see step 2.1.4.). The individual registering the plate will need to check the email address as the plate owner will receive emails regarding imaging as well as plate completed notifications.
- Registering plates
- Log into ISPyB with an appropriate Diamond FedID and select Proposals. Search for the proposal of interest by scrolling or typing the proposal number in the search bar. Select Shipment from the dropdown menu under the proposal number (Figure 3A), which will open the Shipments window with shipments in that proposal. Click +Add shipment on the top right-hand side (Figure 3B) to open the Add New Shipment window, give the shipment a Name, click Automated/Imager, and then click the Add Shipment button at the bottom-left (Figure 3C).
- In the shipment window (Figure 3D), click +Add Container, which will then display the Add container page view (Figure 3E). Choose in the Container type dropdown menu either of the relevant plate types. The page will change to reflect the container type that was chosen. Enter a Barcode and Container name as per email instructions from beamline staff specific to the experimental plates. Note that it is case-sensitive.
- Select VMXi 20 °C imager from the Requested Imager dropdown menu, the Fibonacci imaging schedule from the Imaging Schedule dropdown menu, the crystallization screen from the Crystallization Screen dropdown menu, and the username from the Owner dropdown menu, click the View button, and enter the correct contact email address in the Email box (Figure 3F).
- Enter further details about the plate in the Comments box. Select the relevant sample from the Protein dropdown menu, and use the acronym registered in UAS and approved by Diamond within the experimental proposal. Enter the same name in the Sample Name box; leave the remaining boxes empty.
- Click the +Plate icon to replicate the sample over the whole plate and populate the whole container with green squares. Click +Add Container at the bottom of the page to register the plate. Ask a member of staff at the beamline to transfer the plate to the appropriate imager, where it will be stored and imaged. A visit will be generated when the container is stored in the imagers and the user will receive an email with a link to the plate and its images.
- Viewing imaging results
- Navigate to the proposal of interest (steps 2.1.1), select Containers from the dropdown menu under the proposal number, and observe the list of available containers for the proposal. Select the filter Plates if other sample holder types are present. To narrow the search further, check the My Containers box to only display the most relevant containers associated with the current logged-in user ID. Click the appropriate container by moving the cursor over the individual line and left-clicking the mouse.
- Upon selecting the container, a new view will be presented, showing an overview of the plate (Figure 4A). Click a drop in the plate representation on the left-hand side of the display to show the most recent image from that respective drop. Use arrow keys to navigate between drops, or select individual drops using a mouse/cursor.
- To view historical images of a drop, click on the H button and wait for a popup gallery of images to appear over the current well drop image. Hover the cursor over the individual images to update the main drop image.
- Score images to indicate the status of each drop by pressing the 0 – 9 buttons. To see the individual categories, open the dropdown menu Score at the top-left of the drop image. Look for blue crosses in each of the drop images, which are a result of an algorithm (CHiMP) trained to look for "crystals" within the images.
- Click on the third icon button called Measure at the top right-hand side of the drop image to access a measuring tool. To use this tool, click and drag a line, and a ruler will extend and give the distance in µm.
- To request an additional imaging session, click either Visible or UV from the dropdown box adjacent to the Actions heading towards the bottom of the page. Then, click the Request Plate Imaging button.
- Crystal selection/CHiMP
- To add points for data collection manually, press the +Mark Point button. Hover the cursor over the desired point of interest and select. Wait for a red cross to appear.
NOTE: Up to 100 objects can be created per drop. - When all points are marked, click the +Finish button. Remember to also click the +Finish button before trying to measure objects. To add regions for data collection via grid scans, click the +Mark Region button. Click on the top left-hand point and drag down and right to create a region that will be raster scanned on the beamline. As with points, click the +Finish button when all desired regions are created.
NOTE: It is better to create one larger region than many small regions. - Observe the blue crosses already on the drop images, which are the result of an algorithm designed to automatically locate crystalline objects (CHiMP). To visualize the CHiMP assessment of crystallization drops, click the checkbox Show Auto Scores and then change the dropdown menu for Class. Typically, the most useful setting here is the crystal option (Figure 4B).
NOTE: This is a new feature and it is not guaranteed to find all crystals and may also find other objects that are not crystals. - When all points and regions have been marked in the respective drops, click the Prepare for Data Collection button at the bottom of the page.
- To add points for data collection manually, press the +Mark Point button. Hover the cursor over the desired point of interest and select. Wait for a red cross to appear.
- Preparing samples for data collection
- Observe the list of samples containing the points or regions selected at the previous step, or automatically located (Figure 4C). Add individual points or regions by pressing the + button or add all displayed samples by clicking the Add Current Page To Queue button.
- Filters are available to show only Point, Region, Auto points, or Manual points. To show only those samples that have not been shot (i.e., exposed to X-rays), click the Without Data and Not Completed options above the filter buttons.
- Select individual samples by clicking on the respective line and update the image on the right-hand side of the screen to show the correct drop and individual point. If many samples are in the list, increase the number of samples displayed per page by selecting the dropdown menu with 10 as a default and up to 100 as the maximum number of samples displayed.
- Once all points and regions have been added to the queue, ensure that all experimental data collection parameters are associated with each experiment.
- Use filters for Point, Region, Manual, and Auto. Click on the Point filter and click the Select all checkbox below the filter buttons to simultaneously apply parameters to all samples visible in the current Queued Samples list.
- Select experimental parameters from the dropdown menu on the right-hand side of the screen underneath the drop photo (Figure 4D). For regions, select the option of Grid Scan DMM 10 micron steps, 100 percent transmission. For all other point experiments, select other options from the dropdown menu as appropriate.
- For oscillation data collections, click the Omega Scan DMM 60 degrees 5 percent transmission option to collect the maximum amount of data from an individual sample. Apply small rotations for very small crystals or radiation-sensitive samples and vary the transmission based on prior experience with a particular crystal form. Once all samples have experimental parameters applied correctly, click the Queue Container button at the bottom of the page.
- Once the plate has reached the top of the queue, it will be presented to the beamline, datasets will be collected, and then it will return again to the sample storage within the beamline. Once data collection from a plate is completed, look for an email with a link to follow to access the relevant data.
- Creating sample groups
NOTE: Sample groups may be created to group similar samples across multiple drops or plates. All datasets within these sample groups will be processed using the xia2.multiplex14 pipeline once processed by DIALS. This can be useful when collecting many very small wedges of data and may also be useful to increase signal-to-noise for ligand-binding experiments.- Select Sample Group Management from the dropdown menu under the proposal number. Look for a list of groups if they have already been created by other users. To generate a new group, click the +Create Sample Group button. Click a shipment from the dropdown menu on the Create Sample Group page to see the sample viewer (Figure 5A). Click the container containing the relevant samples from the list that is populated.
- When a container has been clicked, look for a graphic showing the plate overview.
- Click drops individually by clicking on the individual drop (Figure 5B) or click drops in rows or columns by clicking the relevant row letter or column number. When all wells associated with an individual group have been selected, enter a name for the group in the Sample Group Name box, and click the Save Sample Group button. Click the View Sample Groups button on this page to return to the list of already generated sample groups associated with the proposal (Figure 5C).
- Editing sample groups
- Click a sample group from the list of groups on the Sample Group Management page.
- Click the +Edit Sample Group button adjacent to the containers that appear below the group information (Figure 5C).
- Observe the drops, already associated with a sample group, highlighted on the plate overview.
- Add more drops to the sample group by clicking drops, wells, or columns as before.
NOTE: Drops cannot be removed from a sample group. - Once additional drops have been added, edit the Sample Group Name and then save it, or just save it by clicking the Save Sample Group button.
- Visualizing and analyzing the output of sample groups
- Click an individual group from the list of sample groups to display the plate overview of the container or containers associated with the group. The drops included in the group will be highlighted on this display (Figure 5D).
- Look for a list containing the chronological last three multiplex jobs if data have been collected in this group.
- Click the line for a multiplex run to update the processing results below.
- Observe the quick link button, which shows the number of datasets associated with the group. Click this button to open a new Data Collections page displaying the individual dataset collections.
3. Accessing the automatic data processing
NOTE: Once the data have been collected, they are passed through several automatic data processing pipelines. The four standard pipelines used across the MX beamlines at Diamond are also run on data collected at the beamline. They are 'fast_dp', 'xia2 dials', 'xia2 3dii,' and 'autoPROC'15. 'fast_dp' will provide a fast data reduction to quickly assess quality. The other three pipelines will require more computation time and run a variety of different data reduction software packages for comparison. Accordingly, the output is usually of higher quality than the 'fast_dp' output. Datasets collected on the beamline will also run through the automatic multi-crystal merging software 'xia2.multiplex'14, which will merge all datasets within a defined group. Note that while grid scans are not currently automatically processed, the data can be manually processed using the 'xia2.ssx' pipeline. The results of the automatic processing pipelines can be found in ISPyB11 using the following protocol.
- Locating the datasets
- Log into ISPyB as described above and select Proposals.
- Search for the proposal of interest by scrolling or typing the proposal number into the search bar.
- Click the desired visit from the list that appears on the screen to access the Data Collections window for that visit.
- Apply any desired filters.
NOTE: A popular filter is the 'Auto Integrated' filter, which will only display datasets that have run successfully through one or more processing pipelines. This will exclude grid scans as these are not currently automatically processed through ISPyB. - Scroll down the page to find the dataset of interest.
NOTE: Each dataset will display the sample ID, the experimental parameters used, a diffraction image viewer, a crystal image viewer, and a per-image analysis plot for quick observation of data quality.
- To access the auto processing results
- Click the Auto Processing tab underneath the data summary of a specific experiment to inspect the results of the automatic data reduction (Figure 6A).
- Click on the different tabs corresponding to the different pipelines to see a detailed summary of each output.
NOTE: If sample groups have been defined, there will be two tabs corresponding to multiplex jobs. One will correspond to the merging of all datasets in the group up to that point, while the other will correspond to the merging of datasets within that drop only. - Click on the Logs & Files button to download the resulting .mtz files if the processing was successful and any associated log files. Click on the Downstream Processing tab underneath the Auto Processing section to view the output from DIMPLE.
NOTE: DIMPLE will only run if a PDB file was provided during sample submission. - Click on the Logs & Files button to download any resulting output from DIMPLE.
- To access the group multiplex results, open the dropdown menu at the top of the screen with the proposal number written on it and click Sample Group Management. Click on the row corresponding to the desired group within the correct container. Scroll down to find the list of multiplex outputs corresponding to the group as represented visually by a diagram of the plate.
- Click on the desired multiplex output from the list given. Click the xxx Data Sets button, where xxx is the number of merged datasets (Figure 6B).
NOTE: This will open the Data Collections screen, but only the datasets from the selected multiplex job will be shown. - Click on the Auto Processing tab of the top experiment.
- Click on the Multiplex Processing tab that corresponds to the correct number of merged datasets.
- Click on the Logs & Files button to download the .mtz and corresponding log files (as in step 3.2.3.)
- Click on the desired multiplex output from the list given. Click the xxx Data Sets button, where xxx is the number of merged datasets (Figure 6B).
- To access grid scan results
- Navigate to the Data Collections screen for the desired visit. The grid scan data results will be displayed alongside any rotation data collected.
NOTE: There will be no auto processing results. - The image of the crystal drop will have the grid overlaid with a heat map representing the presence of diffraction. Click on a square to view the diffraction image for that position in the grid. Click the dropdown menu at the top of the crystal well image to change what the heat map represents. The default is total intensity of diffraction but can be changed to total spots, estimated resolution, or frames without ice.
- Navigate to the Data Collections screen for the desired visit. The grid scan data results will be displayed alongside any rotation data collected.
4. Data reprocessing
NOTE: Selected datasets can be reprocessed through the ISPyB11 interface using the same processing pipelines that are run automatically with altered settings as defined by the user. A resolution cut-off can be applied; if the symmetry/cell of the crystal is known, then this can also be defined to ensure that the processing pipelines run in the correct setting. Select image ranges across specific datasets can also be merged using available multi-crystal pipelines. This may prove advantageous if systematic radiation damage causes the latter portion of diffraction images to be of poor quality. It is also an option for the user to download their datasets using the protocol described above and run their desired reprocessing software locally, tutorials for which are freely available elsewhere (https://dials.github.io/documentation/tutorials/index.html# ).
- To reprocess multiple individual datasets
- Log into ISPyB and navigate to the datasets of interest (step 3.1).
- Click a dataset and click on the cog icon in the dataset title bar (Figure 6) to open the reprocessing window.
- Configure any desired settings and select which frames will be included in the reprocessing.
NOTE: The image range can be defined either by typing a range in the labeled boxes or by clicking and dragging the desired region in the per-image analysis plot (Figure 7A). - OPTIONAL: To add another dataset for individual reprocessing, click its cog icon and it will appear in the reprocessing window underneath the first dataset. Tick the Process Individually box.
- Click the Integrate button.
- To reprocess multi-crystal data
- Open the reprocessing window from any dataset.
- Click the Multi-Crystal button to open a new screen.
- Scroll down to find a series of per-image analysis plots from experiments during the visit.
- Choose a processing pipeline from the dropdown menu.
- OPTIONAL: Define any resolution limits or known unit cell parameters.
- Click and drag to define image ranges to be included in the multi-crystal reprocessing (Figure 7).
NOTE: This should be done over multiple different plots so that datasets from multiple different crystals are merged. - Click the Integrate button (Figure 7B).
- To access reprocessed data
- Navigate to the Data Collections page for the specific visit (steps 3.1.1-3.1.3).
- Click on the Reprocessing button at the top of the screen.
- Scroll down to locate the desired job.
- Click on the file path in the right-hand column to open the Data Collections screen for the reprocessed data.
- Open the Auto Processing tab and download data as described previously (step 3.2).
NOTE: Any reprocessed jobs are identifiable by the circular arrows symbol next to the pipeline name.
Representative Results
The crystallization facility and VMXi beamline have been used for a wide variety of project types and use cases. Here are a small number of examples to illustrate what users may wish to pursue.
Case study 1: Standard data collection
The beamline enables rapid determination of room-temperature crystal structures from a small number of crystals within a crystallization plate. The minimum number of crystals is dependent on the space group and crystal orientations but is frequently 1-4 although improved data quality may be achieved by merging data from several tens of crystals. A recent example is one of the beamline standards, thaumatin. Multiple crystals, shown in Figure 8A, were marked up for data collection manually as described in protocol section 2.3. These crystals were added to the queue as described in protocol section 2.4 and experimental parameters were selected from the dropdown list. Once experimental parameters had been applied, the plate was queued for data collection. Datasets were collected, automatically scaled, and merged using the xia2.multiplex pipeline as described in protocol section 3. An example output from SynchWeb is shown in Figure 8A middle. Five merged datasets gave rise to a dataset of 1.66 Å resolution. For standard data collection of approximately five crystals in a well, datasets were collected within 2.5 min.
Case study 2: Ligand Binding – Fragment experiment using Mac1 protein
Producing structures of protein-ligand complexes at room temperature may be straightforwardly achieved using the beamline. Ligands may be added to drops on crystallization plates (either manually or by acoustic drop injection) and data measured after a suitable incubation time. In the example described here, a series of fragments were dispensed into wells containing crystals of the SARS-CoV-2 first macrodomain of the protein nsp3 (Mac-1) in a crystallization plate. Two of the wells containing the same fragment were assigned as a group as described in protocol step 2.5. Multiple crystals (42) were marked for data collection as described in protocol steps 2.3 and 2.4, and datasets were collected using standard parameters (60° rotation, 0.1° step, 0.00178 s exposure, 5% transmission, 16 KeV – per crystal) (Figure 8B). Datasets from the two wells were automatically processed using the xia2.dials pipeline and subsequently, the xia2.multiplex pipeline was initiated to automatically merge 22 of these data sets. DIMPLE was then run on the output of these pipelines and yielded maps that clearly showed evidence of the bound fragment. The fragment model was built into the unoccupied density and further refined (Figure 8B right). Room-temperature ligand-bound structures can easily be determined using this series of steps to provide invaluable information and feedback to the structure-based drug design process. For this data collection of 42 crystals across a number of wells, datasets were collected within 10 min.
Case study 3: Structure solution with a low symmetry space group and preferred orientations A stack of multiple crystals with plate-like morphology was produced from crystallization experiments with a c-type gas-binding cytochrome (Figure 8C). By selecting several positions around the edge of the stack where only a single crystal was in the X-ray beam, it was possible to obtain a good quality dataset to 1.75 Å resolution by merging wedges from four crystals, despite a monoclinic (C2) space group. This allowed rapid progress of the project without the need to further optimize the crystallization conditions. This result has been described previously9. For this data collection of four crystals in a well, datasets were collected within 2 min.
Case study 4: Obtaining information and room-temperature structure from microcrystals in a plate using serial crystallography
Often when microcrystals appear in a drop or when users are seeking to optimize micro-crystallization protocols in batch as a precursor to serial crystallography experiments at synchrotron or XFEL sources, it is very helpful to gain rapid feedback on the diffraction properties and unit cell dimensions of different trials using minimal material. In this use case, microcrystals of lysozyme growing in batch were pipetted into a crystallization plate (200 nL volume per drop) and data collected from eight drops using a grid scan with a 10 µm step size (Figure 9). The resulting 25,906 still images were processed using serial crystallography software resulting in a dataset, where 9,891 diffraction patterns were indexed and merged producing a dataset to 2.0 Å resolution that refined well against the published room temperature structure (Rwork = 19.6%, Rfree = 23.6% using PDB 8A9D) (Table 1). This allowed detailed analysis of unit cell distribution and a microcrystal room temperature structure determination that could feed into complex serial crystallography experiments including time-resolved studies. The total volume of microcrystal suspension required was 1.6 µL. For this data collection of microcrystals across eight wells using grid scans, datasets were collected within 40 min.
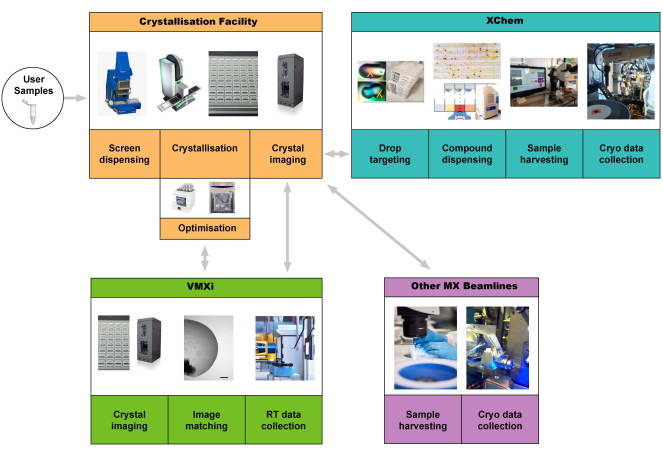
Figure 1: Schematic of the protein-to-structure pipeline integrating crystallization screening, optimization at the crystallization facility, room-temperature automated data collection and processing without sample harvesting at VMXi, XChem fragment screening, and data collection at other MX beamlines. Users can start the pipeline by supplying a sample or by bringing plates to the VMXi beamline. Abbreviation: Versatile Macromolecular Crystallography in situ. Please click here to view a larger version of this figure.
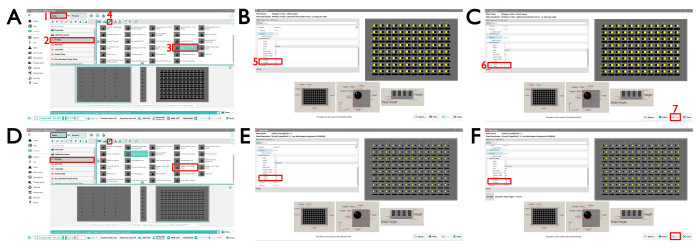
Figure 2: The Mosquito SPT Labtech interface for setting up crystallization plates. (A) (1) The MiTeGen In Situ-1 Setup view. Choose the MiTeGen 2 drop standard plate by going to (2) the 96-well plate type and selecting (3) the MiTeGen plate 2 drop plate. To change the definition parameters for drop 1 and drop 2, which is required for VMXi, click on the (4) edit icon. This opens a new window (B) where (5) the X and Y offsets need to be changed as shown. Select (B) the sub well 2 and (C) sub well 3 and change the values accordingly. (D) The CrystalQuickX Setup view. Choose the CrystalQuickX 2 drop standard plate by going to the 96-well plate type and selecting the MiTeGen plate 2 drop plate. To change the definition parameters for drop 1 and drop 2, which is required for VMXi, click the edit icon the same as above. This opens a new window where (E,F) the X and Y offsets need to be changed as shown. Please click here to view a larger version of this figure.
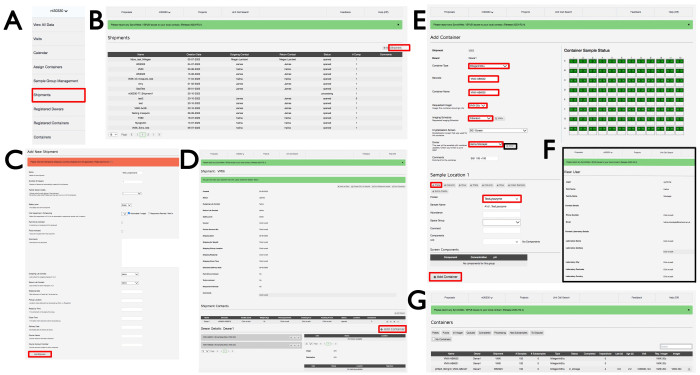
Figure 3: The SynchWeb interface showing how to create a VMXi shipment, register a plate, and check contact details. Screenshots of the various stages of uploading information into the SynchWeb interface are shown from (A) the dropdown menu, (B,C) registering a new shipment, (D) registering a new container, (E) inputting the plate information, (F) check contact details, and (G) a list of registered containers within a proposal. Please click here to view a larger version of this figure.
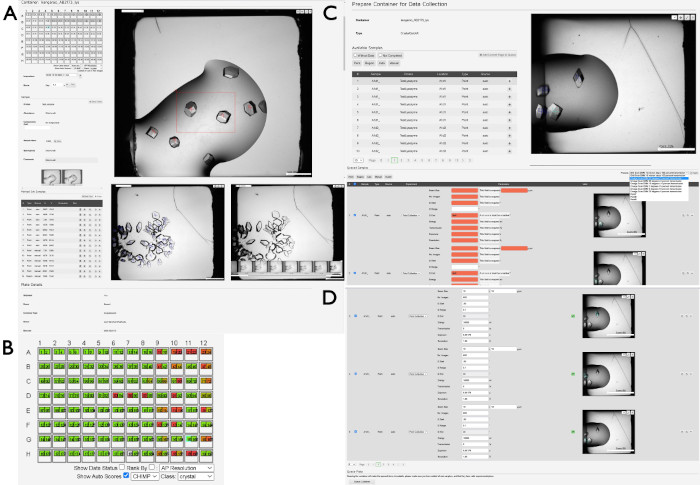
Figure 4: Selecting and preparing samples for data collection using SynchWeb. A series of screenshots showing the various stages of preparing samples for data collection using the SynchWeb interface are displayed. (A) Points and regions of interest are selected from the drop overview. In the lower section of this panel, there is a chronological series of photographs of one drop. (B) An example of the CHiMP output for one plate highlighting results for the 'crystal' category. (C) Adding samples to the queue from the list of selected points and regions and (D) applying parameters for data collection to the queued samples from the dropdown list of beamline-created experiment settings. Note the difference between samples with no experimental parameters (in red) versus those that have correctly applied parameters (top and bottom). At the bottom of this panel is the Queue Container button, which queues the plate to be collected on the beamline. Please click here to view a larger version of this figure.
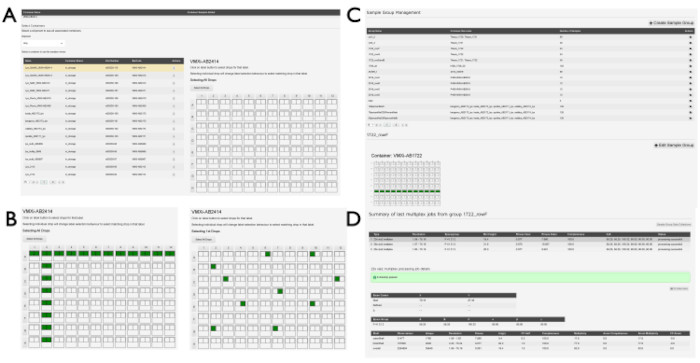
Figure 5: Sample group creation in SynchWeb. A series of screenshots showing the various stages of creating sample groups. (A) The plate(s) containing samples are selected from the relevant shipment and (B) the drops within the plate are selected. These may be individual drops or may be selected by row and/or column. (C) A list of sample groups that have already been created. (D) The outputs of the last three multiplex processing jobs are listed and can be selected to show statistics from the processing pipeline. Please click here to view a larger version of this figure.
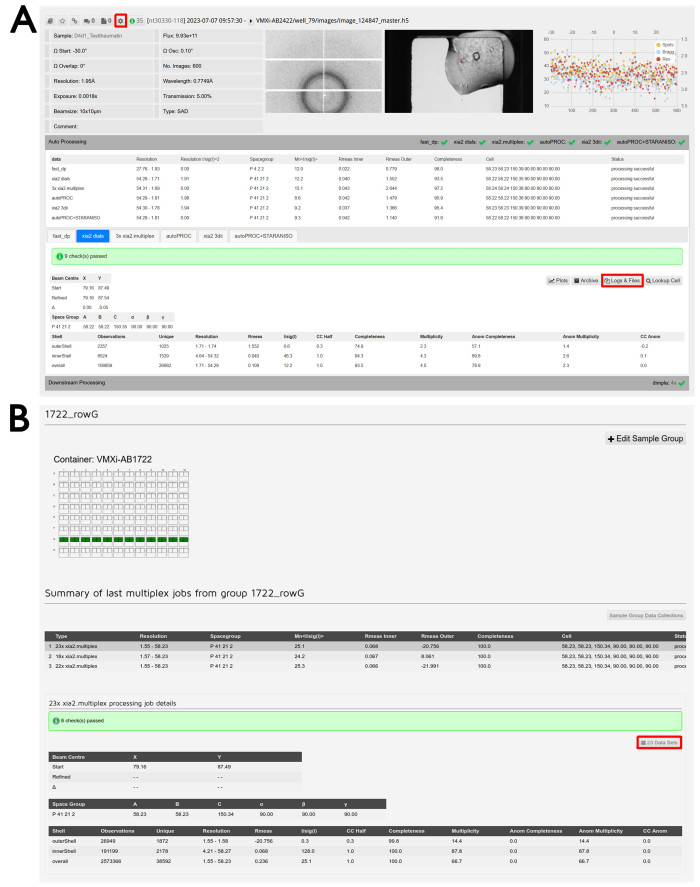
Figure 6: Data processing and data reduction. (A) Screenshot of a processed dataset in ISPyB11. The button to access reprocessing features is highlighted. Sample ID and experimental parameters are shown in the top left and the diffraction image viewer in the middle. Clicking on this image will open an interactive window to examine different images. The crystal image viewer is shown on the right and clicking on this image will also open an interactive window to compare beamline and Formulatrix storage images. Per-image analysis plot is shown on the far right and clicking on this image will open a magnified version of this output. Clicking on the Auto Processing tab will make the autoprocessing visible and make the comparison between the results of the different pipelines easy. Click on the tabs to switch between the different processing pipelines and view the detailed output from the selected pipeline. The Logs & Files button for data download is highlighted. Clicking on the Downstream Processing tab will expand and provide results for any data sets run through post-data reduction pipelines where appropriate. (B) Screenshot from the Sample Group Management screen. The user-defined group name is on the top and the visual description of the included wells can be seen below. A green well indicates that all crystals measured from that drop will be included in the group. A summary of different multiplex jobs performed on that group can be seen and underneath is the detailed output from multiplex. The Data Sets button to examine the included experiments is highlighted. Please click here to view a larger version of this figure.
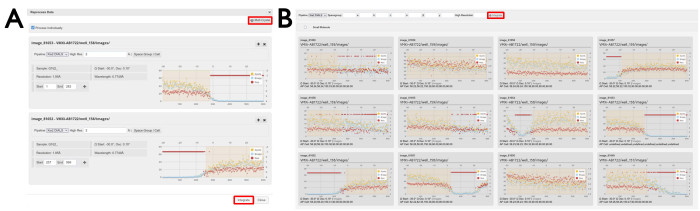
Figure 7: Data reprocessing windows. (A) Individual and (B) multi-crystal datasets. Two individual datasets are displayed where regions of data have been selected. With the Process individually check box ticked, the selected diffraction images will be individually processed by pressing the Integrate button. Clicking the Multi-crystal button will open a display of the individual datasets. To reprocess diffraction images from multiple datasets, regions of images are selected as displayed, and reprocessing is initiated by clicking the Integrate button as highlighted. Please click here to view a larger version of this figure.
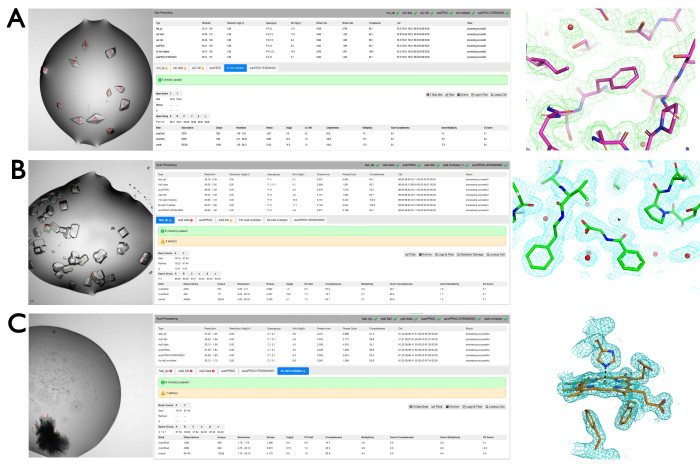
Figure 8: Representative results from the VMXi pipeline. (A) Marked-up crystals for protein thaumatin within a crystallization drop (left panel), data processing results (center panel), and electron density (right panel). (B) Collection on multiple crystals to determine binding of the fragment to SARS-CoV-2 Macro domain. Datasets were collected on multiple crystals in the presence of a fragment from the EU-OPENSCREEN fragment screen using standard experimental settings. Examples of these data collections are shown in this excerpt from SynchWeb. The fragment was built into the corresponding density and further refined as is displayed in the furthest on the right. (C) Marked-up monoclinic crystals in a stack from a challenging crystallization hit used for data collection. Green crosses and red numbers indicate where data were measured using a 10 µm beam and 60° rotation. Four of the resulting wedges were merged to produce a dataset at 1.75 Å resolution. Electron density around the Heme group is displayed on the right. Please click here to view a larger version of this figure.
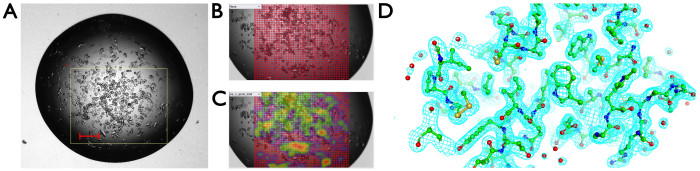
Figure 9: Serial crystallography in the crystallization plate. (A) Optical image of the crystallization drop, with a white box representing the region of interest. (B) Definition of grid scan points. (C) Heat map indicating diffraction. (D) Electron density map arising from a serial crystallography data set from greater than 9,000 still diffraction patterns. Please click here to view a larger version of this figure.
| Resolution (Å) | Completeness (%) | Multiplicity | I/σ(I) | Rsplit | CC1/2 | Unique Observations |
| Overall | 100 | 95.5 | 20.8 | 0.063 | 0.998 | 8422 |
| Low (55.55 – 5.43) | 100 | 147.1 | 81.7 | 0.028 | 0.999 | 488 |
| High (2.03 -2.00) | 100 | 75.3 | 1.2 | 1.092 | 0.410 | 411 |
Table 1: Data statistics for the VMXi RT serial dataset. Abbreviations: I = mean intensity of the scaled observations; Rsplit = a measure of discrepancy of the measured intensities; CC 1/2 = correlation coefficient between two random halves of the dataset.
Discussion
We have described the full procedure from the arrival of a protein sample in the CF to the downloading of the final data by the user for further applications. Critical steps are the production of a high-quality protein sample and appropriate crystal screens, either using commercial sparse matrix screens or optimization screens based on established conditions. This process may take place in the CF, or users may carry out the crystallization procedures in the home laboratories and bring suitable crystallization plates to the beamline. The identification of suitable data collection parameters may be important for certain samples, particularly where radiation damage is a concern. In most cases, automated data processing is fully sufficient to answer the scientific question although users retain the capability to reprocess using the beamline tools, for example, where the space group is ambiguous or only the initial part of the collected data is used to minimize radiation damage effects.
If suitable crystals are not produced from initial crystallization trials, then changes in protein concentration, purity, or crystallization screens may be explored, as may the use of crystal seeding. If crystals do not diffract to a useful resolution at the beamline, then grid scans may be used with an unattenuated beam to assess the inherent diffraction limit and unit cell of the crystals to guide optimization efforts. Crystals that are too small for data collection within plates (e.g., <10 µm) may instead be suitable for serial crystallography or nano focus experiments (e.g., at Diamond beamline VMXm). Solving structures using VMXi data is generally straightforward by molecular replacement, particularly since the advent of Alphafold16 to give effective search models. If this is not successful, crystals may be harvested and cryocooled from plates to enable conventional single-wavelength anomalous diffraction, multiwavelength anomalous diffraction, or long wavelength phasing experiments.
The advantages of this method include the ability to obtain rapid, high-quality datasets and feedback directly from crystallization plates without the need to disturb crystals from the environments in which they have grown. The so-called 'room temperature Renaissance' in structural biology places a premium on structures obtained under non-cryogenic conditions to enable more physiological relevance and protein dynamics to be explored2. Usually, a slightly lower resolution is achieved than for an optimized cryocooled crystal but only when suitable cryo conditions have been established and if the crystals are robust to mechanical handling and opening of the crystallization drop3. A forthcoming application for which this pipeline is very well suited is a large-scale screening of protein-ligand complexes or fragment campaigns at room temperature in drug discovery. Ligands or fragments can either be cocrystallized or added by pipette or acoustic drop ejection prior to room-temperature data collection. Another application is to rapidly measure data from many hundreds or thousands of crystals in a highly efficient manner and then to use the DIALS17 multiplex14 software to extract isomorphous clusters that may represent different biological entities or to establish statistically significant differences between populations of crystals that have been treated in a different way or exposed to different ligands or signal.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We acknowledge the many Diamond Light Source scientists and support team members who contributed to the design, construction, and operation of the VMXi beamline. We are grateful to beamline users, who have contributed ideas later to the development of the crystallization and data collection pipelines. The Crystallization Facility at Harwell is supported by Diamond Light Source Ltd, the Rosalind Franklin Institute, and the Medical Research Council.
Materials
| Formulator | Formulatrix | on request | Liquid handling robot |
| Formulatrix imager | Formulatrix | on request | Crystallisation plate imager |
| Greiner CrystalQuick X | Greiner | Z617644 | Crystallisation plate |
| Gryphon | Art Robbins Instruments | 620-1000-10 | Crystalisation robot |
| MiTeGen Insitu-1 | Mitegen | InSitu-01CL-40 | Crystallisation plate |
| Mosquito LCP | (SPT Labtech) | on request | Crystallisation robot |
| Rock Imager & Maker | Formualtrix | on request |
Software for Imager [1] https://formulatrix.com/protein-crystallization-systems/rock-maker-crystallization-software/ |
| Scorpion | Art Robbins Instruments | 640-1000-10 |
Liquid handling robot https://www.artrobbins.com/scorpion |
References
- Lynch, M. L., Snell, M. E., Potter, S. A., Snell, E. H., Bowman, S. E. J. 20 years of crystal hits: Progress and promise in ultrahigh-throughput crystallization screening. Acta Crystallographica Section D Structural Biology. 79 (Pt 3), 198-205 (2023).
- Fischer, M. Macromolecular room temperature crystallography. Quarterly Reviews of Biophysics. 54, 1 (2021).
- Helliwell, J. R. What is the structural chemistry of the living organism at its temperature and pressure. Acta Crystallographica Section D Structural Biology. 76 (Pt 2), 87-93 (2020).
- Thorne, R. E. Determining biomolecular structures near room temperature using x-ray crystallography: Concepts, methods and future optimization. Acta Crystallographica Section D Structural Biology. 79 (Pt 1), 78-94 (2023).
- Keedy, D. A., et al. Crystal cryocooling distorts conformational heterogeneity in a model michaelis complex of dhfr. Structure. 22 (6), 899-910 (2014).
- Huang, C. Y., et al. Probing ligand binding of endothiapepsin by ‘temperature-resolved’ macromolecular crystallography. Acta Crystallographica Section D Structural Biology. 78 (Pt 8), 964-974 (2022).
- Sanchez-Weatherby, J., et al. Vmxi: A fully automated, fully remote, high-flux in situ macromolecular crystallography beamline. Journal of Synchrotron Radiation. 26 (Pt 1), 291-301 (2019).
- Jacquamet, L., et al. Automated analysis of vapor diffusion crystallization drops with an x-ray beam. Structure. 12 (7), 1219-1225 (2004).
- Mikolajek, H., et al. Protein-to-structure pipeline for ambient-temperature in situ crystallography at vmxi. IUCrJ. 10, 420-429 (2023).
- Douangamath, A., et al. Achieving efficient fragment screening at xchem facility at diamond light source. Journal of Visualised Experiments. (171), (2021).
- Delageniere, S., et al. Ispyb: An information management system for synchrotron macromolecular crystallography. Bioinformatics. 27 (22), 3186-3192 (2011).
- Fisher, S. J., Levik, K. E., Williams, M. A., Ashton, A. W., Mcauley, K. E. Synchweb: A modern interface for ispyb. Journal of Applied Crystallography. 48 (Pt 3), 927-932 (2015).
- Jenkins, J., Cook, M. Mosquito®: An accurate nanoliter dispensing technology. JALA: Journal of the Association for Laboratory Automation. 9 (4), 257-261 (2016).
- Gildea, R. J., et al. Xia2.Multiplex: A multi-crystal data-analysis pipeline. Acta Crystallographica Section D Structural Biology. 78 (Pt 6), 752-769 (2022).
- Winter, G., Mcauley, K. E. Automated data collection for macromolecular crystallography. Methods. 55 (1), 81-93 (2011).
- Jumper, J., et al. Highly accurate protein structure prediction with alphafold. Nature. 596 (7873), 583-589 (2021).
- Winter, G., et al. Dials as a toolkit. Protein Science. 31 (1), 232-250 (2022).

