Cell-Based AAV Neutralizing Antibody Assay: A Colorimetric Technique to Detect Neutralizing Antibodies Against Specific Adeno-Associated Viruses in Serum
Abstract
Source: Bass-Stringer, S., et al. A Step-By-Step Method to Detect Neutralizing Antibodies Against AAV using a Colorimetric Cell-Based Assay. J. Vis. Exp. (2021)
In this video, we demonstrate an in vitro colorimetric assay to detect the presence of neutralizing antibodies against alkaline phosphatase-expressing recombinant adeno-associated virus serotype 6 (rAAV6) vectors in sheep serum samples. The assay utilizes the reaction between the AAV encoding a human placental alkaline phosphatase gene and its substrate to generate an insoluble purple product.
Protocol
All procedures involving animal models have been reviewed by the local institutional animal care committee and the JoVE veterinary review board.
A schematic overview of the assay protocol is provided in Figure 1.
1. Initial preparation
- For assessment in sheep: collect blood in 8 mL serum separator clot activator tubes (see Table of Materials), leave the blood sample at room temperature (RT) for 20-30 min, and subsequently, spin down at 2,100 x g for 15 min. The clear supernatant that forms at the top of the tubes is serum. Aliquot the clear aqueous phase into microcentrifuge tubes and store at -80 °C.
NOTE: The serum at -80 °C remains stable for ~5 years. Blood was collected from the carotid vein using a 16 G needle (tip cut-off) and syringe from conscious animals. - Heat inactivated fetal bovine serum (FBS) by placing it in a water bath at 56 °C for 30 min, and swirl intermittently. For precision, place a thermometer in a second bottle containing an equivalent volume of water and add it to the heat bath at the same time as the FBS bottle. Begin timing when the thermometer reaches 56 °C.
- Employ proper aseptic technique and cell culture practice for all subsequent steps performed in the cell culture hood. Spray 70% ethanol on all objects and the hood before use and clean with 1% sodium hypochlorite upon completion.
- Make complete Dulbecco's Modified Eagle Medium (DMEM) by combining high glucose (4.5 g/L) DMEM (89%) with heat-inactivated FBS (10%) and Penicillin Streptomycin (1%). Combine and filter using a sterile vacuum filtration system (0.22 μm pore size, polyethersulfone membrane) (see Table of Materials). Store complete DMEM wrapped in foil at 4 °C.
- Establish HT1080 cells (see Table of Materials) and passage in a 75 cm2 square flask. Create multiple frozen stocks of cells. Do not use cells after 20 passages as further passaging may influence the assay results.
2. Day 1 – Plating of cells
- Passage HT1080 cells when they reach ~80% confluency.
- Pre-warm complete DMEM (prepared in step 1.4), 0.05% trypsin-EDTA, and 1x phosphate-buffered saline (PBS) to 37 °C in a water bath. Remove the growth medium from passaged cells using an aspiration system.
NOTE: All aspirations in this protocol use a vacuum system with a tube attached to a sterile 5 mL serological pipette. - Wash the cells in 10 mL of pre-warmed (37 °C) 1x PBS and trypsinize cells for 3-4 min in 4 mL of pre-warmed 0.05% trypsin-EDTA to detach the cells from the flask.
- Inactivate the trypsin by adding 6 mL of pre-warmed complete DMEM and pipette the cells into a 50 mL tube. Calculate the number and concentration of viable cells using a hemocytometer and the trypan blue exclusion method.
- Dilute the cells to a concentration of 1 x 105 cells/mL in pre-warmed complete DMEM. Seed 100 μL of cells/well into clear 96-well flat-bottomed plates (1 x 104 cells per well). Incubate the plate at 37 °C, 5% carbon dioxide (CO2) overnight for 16-22 h.
3. Day 2 – Infecting the cells
- Remove plate/s from the incubator and use a light microscope to confirm that cells are evenly dispersed within the wells and that the confluency is ~50%. If cells are not within a range of 45%-55% confluency, repeat the 'Day 1' protocol and adjust initial cell concentration accordingly.
- Generate serial dilutions of the serum samples of interest in 1.5 mL microcentrifuge tubes using prewarmed complete DMEM as the diluent. Table 1 demonstrates the generation of a dilution cascade for triplicate samples.
- To perform the assay in triplicate, prepare a 7.5 x106 vector genomes (vg)/μL of working solution of AAV6-hPLAP (see Table of Materials) by diluting a virus stock solution in 1x PBS.
- Add 66 μL of the 7.5 x 106 vg/μL virus working solution to each tube containing 264 μL of serum/media dilution (330 μL of total volume/dilution, see Table 1).
NOTE: This is a robust assay that does not require perfect culture conditions. However, to accurately quantitate and ensure each assay run is reliable, it is necessary to include the following: (1) a virus and media-only control, (2) a media-only control, and (3) a NAb positive control sample on all plates under the same experimental conditions. The volume described (330 μL) accounts for triplicate samples+10% of the serum and virus mixture. Performing replicates is highly recommended for the accurate determination of neutralizing activity.
- Mix the virus/serum dilutions by pipetting and place the tubes containing the virus/serum mixtures in an incubator at 37 °C, 5% CO2 for 30 min to allow potential neutralization to occur.
- Pipette 100 μL of the virus/serum mixture to each well on the 96-well plate containing 1 x 104 cells/well for each dilution.
NOTE: This will generate a final viral concentration of 15k viruses/cell multiplicity of infection (MOI) in each well. Table 2 provides an example 96-well sample plate layout for assessing samples to a 1/512 dilution. - Wrap the 96-well plate containing cells, serum, and AAV-hPLAP in foil and place in an incubator at 37 °C, 5% CO2 overnight for 16-24 h to allow AAV entry into the cells.
4. Day 3 – Fixing and adding substrate to the cells
- Pre-warm an aliquot of 1x PBS to 37 °C (~25 mL/96-well plate). Cool separate aliquots of PBS (~25 mL/96-well plate) and double-distilled H2O (DDW, ~25 mL/96-well plate) to 4 °C. Dissolve a pellet of BCIP/NBT (see Table of Materials) in 10 mL of DDW in a 50 mL conical centrifuge tube by vortexing (10 mL is enough for 2 x 96 well plates).
- Aspirate the media from the wells of the 96-well plate using a serological pipette or similar attached to a suction-based aspiration system or fume hood vacuum. Gently place the tip of the serological pipette into the well and remove the media taking caution not to disrupt the adhered cells.
- Add 50 μL of RT 4% PFA to each well using a pipette. Wrap the plate in foil and leave it at RT for 10 min to fix the cells.
CAUTION: Paraformaldehyde (PFA) is a probable carcinogen and is toxic from skin or eye contact or inhalation. Handle in a fume hood with proper personal protective equipment as well as a facemask. Make fresh 4% PFA diluted in PBS (~7 mL required per 96-well plate).
- Add 50 μL of RT 4% PFA to each well using a pipette. Wrap the plate in foil and leave it at RT for 10 min to fix the cells.
- Wash and aspirate the cells with 200 μL of RT 1x PBS. Repeat this step twice.
NOTE: A multichannel pipette is an efficient option for the pipetting steps. - Pipette 200 μL of pre-warmed PBS into each well, wrap the plate in foil, and incubate at 65 °C for 90 min to denature endogenous alkaline phosphatase activity.
- Aspirate wells and wash cells with 200 μL of cold (4 °C) PBS. Aspirate again, wash in 200 μl of cold DDW, and aspirate again.
- Pipette 50 μL of the dissolved BCIP/NBT (prepared in step 4.1) into each well.
- Wrap the plate in foil and incubate at RT for 2-24 h.
NOTE: Be consistent with incubation time between runs; the time flexibility allows users to photograph wells either on day 3 or the following day. - Using a light microscope camera, take photos of each well using a 4x objective lens, ensuring the same exposure, white balancing, and light settings are used consistently for all assays performed.
- Position each well identically and ensure the edges of the well are not visible in the photos. Save photos in TIF format or similar.
NOTE: Specific settings will vary between microscopes, but quantitation will be most effective if the background lighting is high and consistent throughout the wells (Figure 1B).
- Position each well identically and ensure the edges of the well are not visible in the photos. Save photos in TIF format or similar.
Table 1: Volumes of serum and diluent required to generate serial dilutions of serum in triplicate.
| Dilution cascade label | Dilution | 3 x sample (240 μL) + 10% buffer volume (24 μL) | Ratio of serum:media |
| Dilution 1 (D1) | 1/2 | 264 μL serum 264 μL media | 50:50 |
| Dilution 2 (D2) | 1/4 | 264 μL D1 + 264 μL media | 25:75 |
| Dilution 3 (D3) | 1/8 | 264 μL D2 +264μL media | 12.5:87.5 |
| Dilution 4 (D4) | 1/16 | 264 μL D3 +264 μL media | 6.25:93.75 |
| Dilution 5 (D5) | 1/32 | 264 μL D4 +264 μL media | 3.13:96.87 |
| Dilution 6 (D6) | 1/64 | 264 μL D5 +264 μL media | 1.56:98.44 |
| Dilution 7 (D7) | 1/128 | 264 μL D5 +264 μL media | 0.78:99.22 |
| Dilution 8 (D8) | 1/256 | 264 μL D5 +264 μL media | 0.39:99.61 |
| Dilution 9 (D9) | 1/512 | 264 μL D7 + 264 μL media | 0.2:99.8 |
| Dilution 10 (D10) | 1/2048 | 132 μL D8 + 396 μL media | 0.05:99.95 |
| Dilution 11 (D11) | 1/8192 | 132 μL D9 + 396 μL media | 0.01:99.99 |
| Dilution 12 (D12) | 1/32768 | 132 μL D10 + 396 μL media | 0.003:99.997 |
Table 2: Example 96-well plate layout for assessing naïve serum samples in dilutions ranging from 1/2 to 1/512. Higher dilutions are incorporated into the assay if assessing a sample known to be positive for AAV NAbs (post-administration samples) or if a higher titer is required. MO (-C): Media-only control. VO (+C): Virus and media-only control. mAb: Monoclonal antibody against AAV (NAb positive control).
| Serum sample #1 | Serum sample #2 | Serum sample #3 | Mono AB (mAB), controls and extra samples | |||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | 1/2 | 1/2 | 1/2 | 1/2 | 1/2 | 1/2 | 1/2 | 1/2 | 1/2 | 50 ng MAb |
50 ng MAb |
50 ng MAb |
| B | 1/4 | 1/4 | 1/4 | 1/4 | 1/4 | 1/4 | 1/4 | 1/4 | 1/4 | 5 ng MAb |
5 ng MAb |
5 ng MAb |
| C | 1/8 | 1/8 | 1/8 | 1/8 | 1/8 | 1/8 | 1/8 | 1/8 | 1/8 | 0.5 ng MAb |
0.5 ng MAb |
0.5 ng MAb |
| D | 1/16 | 1/16 | 1/16 | 1/16 | 1/16 | 1/16 | 1/16 | 1/16 | 1/16 | MO (-C) | MO (-C) | MO (-C) |
| E | 1/32 | 1/32 | 1/32 | 1/32 | 1/32 | 1/32 | 1/32 | 1/32 | 1/32 | VO (+C) | VO (+C) | VO (+C) |
| F | 1/64 | 1/64 | 1/64 | 1/64 | 1/64 | 1/64 | 1/64 | 1/64 | 1/64 | Sample #1 1/512 |
Sample #1 1/512 |
Sample #1 1/512 |
| G | 1/256 | 1/256 | 1/256 | 1/256 | 1/256 | 1/256 | 1/256 | 1/256 | 1/256 | Sample #2 1/512 |
Sample #2 1/512 |
Sample #2 1/512 |
| H | 1/512 | 1/512 | 1/512 | 1/512 | 1/512 | 1/512 | 1/512 | 1/512 | 1/512 | Sample #3 1/512 |
Sample #3 1/512 |
Sample >#3 1/512 |
Representative Results
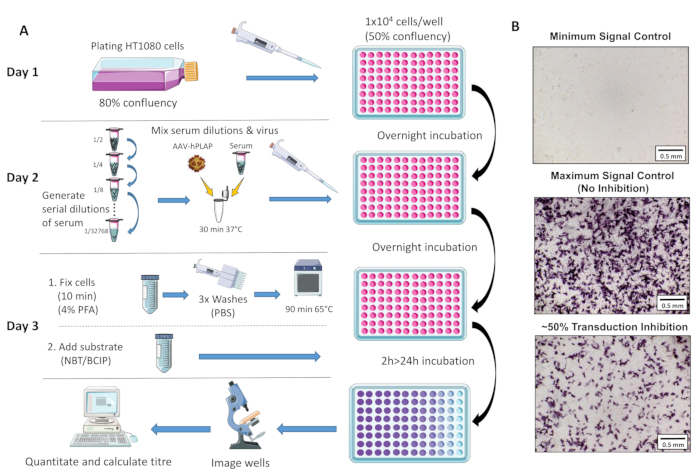
Figure 1: Schematic diagram of NAb assay protocol. (A) Visual representation of the NAb assay illustrating the primary steps involved in the three-day protocol. Briefly, cells are grown and plated overnight. The following day, serial dilutions of serum are prepared, incubated with AAV, and then incubated with the cells overnight. The next day, cells are fixed, washed, incubated, combined with the substrate, and incubated again, followed by imaging and quantitation. (B) Representative images of a minimum signal control (complete AAV inhibition), a maximum signal control (no inhibition), and an ovine serum sample with ~50% signal inhibition. Scale bar = 0.5 mm.
Declarações
The authors have nothing to disclose.
Materials
| 0.05% Trypsin/EDTA | Gibco | 25300-054 | |
| 50 mL conical centrifuge tube | Falcon | 14-432-22 | Or equivalent |
| 75 cm2 square flasks | Falcon | 353136 | Or equivalent |
| 96 well flat bottomed plate | Falcon | 353072 | |
| AAV6-CMV-hPLAP Vector | Muscle Research & Therapeutics Lab (University of Melbourne, Australia) AAV6-CMV-hPLAP can be provided upon request. | ||
| Aluminium foil | |||
| Anti-AAV6 (intact particle) mouse monoclonal antibody, (ADK6) | PROGEN | 610159 | Positive control monoclonal antibody |
| BCIP/NBT | SIGMAFAST | B5655 | |
| Cell and tissue culture safety cabinet | |||
| Electronic Pipette | 5 & 10 mL stripette inserts | ||
| Fetal Bovine Serum | Gibco | 10099-141 | |
| Haemocytometer | |||
| High glucose Dulbecco's Modified Eagle Medium (DMEM) | Gibco | 11965118 | |
| HT1080 cells | ATCC | ||
| Incubator | 37 °C, 5% CO2 | ||
| Light microscope with camera | Capable of taking photos with a 4x objective lens | ||
| Oven | For a 65 °C incubation | ||
| Paraformaldehyde | MERCK | 30525-89-4 | |
| Water bath | |||
| Penicillin Streptomycin | Gibco | 15140-122 |

