A 1.5 Hour Procedure for Identification of Enterococcus Species Directly from Blood Cultures
PREPARAÇÃO DO INSTRUTOR
CONCEITOS
PROTOCOLO DO ALUNO
1. Specimen Collection and Preparation
- Collect venous blood from patients suspected of sepsis and put into two BACTEC (BD, Sparks, MD) blood culture bottles, one aerobic and one anaerobic, together comprising one blood culture set.
- Place bottles into the BACTEC 9240 instrument at 35°C.
- Incubate until the instrument is triggered into alarm due to growth of organisms in the blood culture bottle.
- Remove bottle from the blood culture instrument.
2. Preparation of Reagents Prior to Staining
- Prepare the wash solution by adding 4 mL of 60x Wash Solution followed by 240 mL of fresh deionized or distilled water directly to the Staining Dish.
- Put the staining dish in the 55°C water bath to warm.
- Store the remaining concentrate at 2-8°C.
- Remove the Mounting Medium from the refrigerator and warm to room temperature.
3. Gram Staining
- Gently swirl the blood culture bottle.
- Using a needle and syringe or a subculturing apparatus, remove blood (approximately 5 mL) from the bottle and place in a sterile screw capped tube.
- Using a sterile pipette, place one large drop of blood onto a glass microscope slide.
- Air dry the slide and fix in methanol for 10 seconds.
- Allow slide to air dry.
- Perform a Gram stain on the blood film to determine the morphology of the organism growing in the blood culture bottle.
- Using an oil immersion lens, view slide. Gram positive cocci in pairs and short chains observed on the Gram stain is most consistent with enterococcus species.
- This Gram stain result then drives the selection of the proper PNA FISH probe kit to use for bacterial identification. This blood culture will be stained using the E. faecalis/OE PNA FISH stain.
4. PNA FISH Stain
- Mix the tube containing the blood gently by swirling prior to smear preparation.
- Place one drop of Fixation Solution on the well of a PNA FISH Microscope Slide.
- Transfer 10 μL or one small drop from the tube with the positive blood culture to the Fixation Solution and mix gently to emulsify.
- Fix the smears by heating them for 20 min. at 55°C on a heating plate.
5. Quality Control Material
One positive and one negative quality control slide must be tested with each batch of slides for staining.
- Use E. faecalis/OE Control Slides purchased from AdvanDx for this purpose or prepare smears from liquid cultures of reference strains of E. faecalis and E. faecium as Positive Control either on separate slides or mixed on one slide and Staphylococcus spp or Streptococcus spp as Negative Control material.
The QC results should be able to monitor for appropriate testing conditions, particularly those affecting hybridization stringency and cell wall penetration, since PNA methodology is designed to optimize cell wall penetration
6. Hybridization
- Add one drop of E. faecalis/OE PNA to the well on the microscope slide with the smear.
- Add coverslip. Avoid air bubbles.
- Incubate for 30 ± 5 min. at 55 ± 1°C on heating plate
- Following incubation place the slides in slide carrier
7. Stringent Wash
- Immerse slide carrier in the preheated Wash Solution at 55°C and carefully remove the coverslip. Often, the coverslip slides off by gently agitating the slide in the Wash Solution. Occasionally, the coverslip must be gently pushed off with forceps.
- Incubate in the wash solution for 30 ± 5 min. at 55 ± 1°C.
- Remove the slides from the wash solution
- Allow the slide to air dry
8. Mounting
- Add one drop of Mounting Medium to each slide
- Add coverslip. Avoid air bubbles.
- Examine the slide on a fluorescence microscope within 2 hours.
- Do not expose the slides to direct sun light or other strong light sources as this may lead to fluorescence quenching.
9. Interpretation of Results
The fluorescence microscope used for slide examination must be equipped with the AdvanDx Dual Band Filter and a 60x or 100x oil objective. The QC slides should be examined first to confirm that hybridization did occur. E. faecalis should appear as bright green fluorescent cocci in multiple fields of view and the E. faecium will appear as bright red cocci. Non-enterococci control slide should appear nonfluorescent. After confirming the system in controls, the patient slides can be examined. At first the blood film will appear reddish but the bright red and green cocci will be quite apparent.
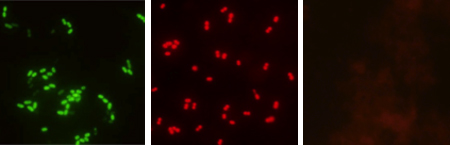
Figure 1.Representative examples of green-positive E. faecalis (left), red positive E. faecium (middle), and negative (right) test results.
10. Representative Results
Three institutions were included in a multi-center clinical trial evaluating this PNA FISH stain and comparing a 2.5 hour protocol to the shortened 1.5 hour protocol that has just been reviewed. A total of 152 routine Gram positive cocci in pairs and chains (GPC) positive blood culture bottles were included in the studies. There was 100% (152/152) agreement between results of the modified and the original assay procedure for E. faecalis/OE PNA FISH: 41/41 E. faecalis; 33/33 other enterococci and 78/78 other GPC (Table 1). This staining method had exquisite sensitivity and specificity during this clinical trial (Table 2).
| Routine Methods | E. faecalis /OE PNA FISH Standard Procedure | E. faecalis/OE PNA FISH Short Procedure | |
| E. faecalis | 41 | 41 (Green Positive) | 41 (Green Positive) |
| E. faecium | 27 | 27 (Red Positive) | 27 (Red Positive) |
| E. casseliflavus | 2 | 2 (Red Positive) | 2 (Red Positive) |
| E. gallinarum | 2 | 2 (Red Positive) | 2 (Red Positive) |
| Outro Enterococcus spp. | 2 | 2 (Red Positive) | 2 (Red Positive) |
| S. pneumoniae | 17 | 17 (Negative) | 17 (Negative) |
| S. viridans | 33 | 33 (Negative) | 33 (Negative) |
| S. mitis | 1 | 1 (Negative) | 1 (Negative) |
| S. pyogenes | 3 | 3 (Negative) | 3 (Negative) |
| S. bovis | 2 | 2 (Negative) | 2 (Negative) |
| S. salivarius | 1 | 1 (Negative) | 1 (Negative) |
| S. sanguinis | 2 | 2 (Negative) | 2 (Negative) |
| S. agalagtae | 7 | 7 (Negative) | 7 (Negative) |
| Outro Streptococcus spp. | 7 | 7 (Negative) | 7 (Negative) |
| Abiotrophia spp. | 3 | 3 (Negative) | 3 (Negative) |
| Peptostrepococcus spp. | 2 | 2 (Negative) | 2 (Negative) |
| Total | 152 | 152/152 100% Agreement |
152/152 100% Agreement |
Table 1. Listing of bacteria tested using both the standard (2.5hr) and rapid (1.5 hr) protocol.
| Sensitivity E.faecalis | Sensitivity Other Enterococcus spp. | Specificity |
| 100% (41/41) 95% CI (93.0-100) |
100% (33/33) 33/33 95% CI (91.3-100) |
100% (78/78) 95% CI (96.2-100) |
Table 2. Sensitivity and Specificity of the 1.5 hour PNA FISH protocol.
A 1.5 Hour Procedure for Identification of Enterococcus Species Directly from Blood Cultures
Learning Objectives
List of Materials
Reagents
E. faecalis/OE PNA FISH is comprised of the following kit components:
- Fixation Solution Fixation Solution 3 mL phosphate-buffered saline with detergent.
- E. faecalis/OE PNA E. faecalis/OE PNA 1.5 mL PNA probes in hybridization. solution. Contains 30% formamide.
- 60x Wash Solution 60x Wash Solution 50 mL Tris-buffered saline with detergent
- Mounting Medium Mounting Medium 3 mL photobleaching inhibitor in Glycerol
- Quality control slides
- AdvanDX
Preparação do Laboratório
Enterococci are a common cause of bacteremia with E. faecalis being the predominant species followed by E. faecium. Because resistance to ampicillin and vancomycin in E. faecalis is still uncommon compared to resistance in E. faecium, the development of rapid tests allowing differentiation between enterococcal species is important for appropriate therapy and resistance surveillance. The E. faecalis OE PNA FISH assay (AdvanDx, Woburn, MA) uses species-specific peptide nucleic acid (PNA) probes in a fluorescence in situ hybridization format and offers a time to results of 1.5 hours and the potential of providing important information for species-specific treatment. Multicenter studies were performed to assess the performance of the 1.5 hour E. faecalis/OE PNA FISH procedure compared to the original 2.5 hour assay procedure and to standard bacteriology methods for the identification of enterococci directly from a positive blood culture bottle.
Enterococci are a common cause of bacteremia with E. faecalis being the predominant species followed by E. faecium. Because resistance to ampicillin and vancomycin in E. faecalis is still uncommon compared to resistance in E. faecium, the development of rapid tests allowing differentiation between enterococcal species is important for appropriate therapy and resistance surveillance. The E. faecalis OE PNA FISH assay (AdvanDx, Woburn, MA) uses species-specific peptide nucleic acid (PNA) probes in a fluorescence in situ hybridization format and offers a time to results of 1.5 hours and the potential of providing important information for species-specific treatment. Multicenter studies were performed to assess the performance of the 1.5 hour E. faecalis/OE PNA FISH procedure compared to the original 2.5 hour assay procedure and to standard bacteriology methods for the identification of enterococci directly from a positive blood culture bottle.
Procedimento
Enterococci are a common cause of bacteremia with E. faecalis being the predominant species followed by E. faecium. Because resistance to ampicillin and vancomycin in E. faecalis is still uncommon compared to resistance in E. faecium, the development of rapid tests allowing differentiation between enterococcal species is important for appropriate therapy and resistance surveillance. The E. faecalis OE PNA FISH assay (AdvanDx, Woburn, MA) uses species-specific peptide nucleic acid (PNA) probes in a fluorescence in situ hybridization format and offers a time to results of 1.5 hours and the potential of providing important information for species-specific treatment. Multicenter studies were performed to assess the performance of the 1.5 hour E. faecalis/OE PNA FISH procedure compared to the original 2.5 hour assay procedure and to standard bacteriology methods for the identification of enterococci directly from a positive blood culture bottle.
