Establishment of Epstein-Barr Virus Growth-transformed Lymphoblastoid Cell Lines
Summary
We describe a method for generating transformed B cell lines using Epstein-Barr virus. We also illustrate a novel assay that can identify B cells destined to undergo transformation as early as three days after infection.
Abstract
Infection of B cells with Epstein-Barr virus (EBV) leads to proliferation and subsequent immortalization, resulting in establishment of lymphoblastoid cell lines (LCL) in vitro. Since LCL are latently infected with EBV, they provide a model system to investigate EBV latency and virus-driven B cell proliferation and tumorigenesis1. LCL have been used to present antigens in a variety of immunologic assays2, 3. In addition, LCL can be used to generate human monoclonal antibodies4, 5 and provide a potentially unlimited source when access to primary biologic materials is limited6, 7.
A variety of methods have been described to generate LCL. Earlier methods have included the use of mitogens such as phytohemagglutinin, lipopolysaccharide8, and pokeweed mitogen9 to increase the efficiency of EBV-mediated immortalization. More recently, others have used immunosuppressive agents such as cyclosporin A to inhibit T cell-mediated killing of infected B cells7, 10-12.
The considerable length of time from EBV infection to establishment of cell lines drives the requirement for quicker and more reliable methods for EBV-driven B cell growth transformation. Using a combination of high titer EBV and an immunosuppressive agent, we are able to consistently infect, transform, and generate LCL from B cells in peripheral blood. This method uses a small amount of peripheral blood mononuclear cells that are infected in vitroclusters of cells can be demonstrated. The presence of CD23 with EBV in the presence of FK506, a T cell immunosuppressant. Traditionally, outgrowth of proliferating B cells is monitored by visualization of microscopic clusters of cells about a week after infection with EBV. Clumps of LCL can be seen by the naked eye after several weeks. We describe an assay to determine early if EBV-mediated growth transformation is successful even before microscopic clusters of cells can be demonstrated. The presence of CD23hiCD58+ cells observed as early as three days post-infection indicates a successful outcome.
Protocol
1. EBV stock preparation
- Subculture exponentially growing B95-8 cells (ATCC # CRL 1612; 13) at 3 x 105 cells/ml in a 75cm2 tissue culture flask using sterile technique. Cells are grown in complete RPMI 1640 containing 10% heat-inactivated fetal bovine serum (FBS), Penicillin/Streptomycin at 100U/ml and 100 μg/ml, respectively, and Amphotericin B at 0.5 μg/ml at 37°C in the presence of 5% CO2.
- Forty-eight hours later, resuspend cells in fresh complete RPMI 1640 at 1 x 106 cells/ml. To induce virus production, stimulate cells with 20ng/ml tetradecanoyl phorbol acetate (TPA) for 1 hour in a standard CO2 incubator. Wash cells three times with RPMI 1640 to remove TPA.
- Resuspend cells in the original volume of complete RPMI 1640 (from 1.2) and place the flask in CO2 incubator for 96 hours. This method has been shown to produce high titers of infectious virus particles6.
- Centrifuge at 600 x g for 10 min at 4°C to separate EBV-containing culture supernatant from cells. Filter supernatant through a 0.45-micron filter, aliquot, and store at -70°C for over a year.
An alternative is to obtain the supernatant from B95-8 cells containing EBV from the ATCC (VR-1492) and use at the dilution recommended by the ATCC.
2. Isolation of peripheral blood mononuclear cells (PBMC)
- Draw 10 ml blood from donor into a heparinized syringe or a heparinized blood tube. Dilute blood with 20 ml PBS at room temperature in a 50 ml conical tube.
- Underlay diluted blood with 15 ml Ficoll Hypaque lymphocyte separation medium. Centrifuge at 225 x g without brake at room temperature for 30 minutes.
- Remove buffy coat and transfer into a new 50 ml conical tube. Raise volume to 50 ml with PBS and spin at 600 x g at room temperature for 10 minutes.
- Pour off supernatant. Wash cells by resuspending pellet in 50 ml PBS and spinning at 600 x g at room temperature for 10 minutes. Wash twice more in a similar manner.
- Resuspend washed cells in 1 ml of complete RPMI. Use 5 μL of cells to prepare a 1/10 dilution in Trypan blue. Count live cells using a hemocytometer.
- Adjust the volume using complete RPMI in a 25 cm2 tissue culture flask to obtain a cell concentration of 2 x 106/ml.
3. Infection with EBV
- Add FK506 (AG Scientific) to the cell suspension from 2.6 to a final concentration of 20 nM. Place flask in CO2 incubator at 37°C for one hour.
- Rapidly thaw an aliquot of EBV. Remove flask from incubator and add EBV to the cells at 1/10 dilution. Typically, this dilution provides an MOI of 50-100. Swirl flask to mix and place it upright in CO2 incubator at 37°C.
Note that Step 4, performed to predict successful outcome, is an optional step. If Step 4 is to be performed, you will need to incubate an additional flask of un-infected PBMC at 2 x 106 cells/ml as control.
4. Identifying proliferating population of cells (Optional step)
- Prepare FACS buffer: PBS + 5% FBS. Store at 4°C.
- Make the following antibody mixes in 50 μL volume each for staining cells in Step 4.6. Antibody dilutions should be made in FACS buffer containing 1mg/ml of mouse IgG (Sigma) to inhibit non-specific binding by antibodies.
- single-stained with PE conjugated anti-CD23 antibody (1/50 dilution)
- single-stained with FITC conjugated anti-CD58 antibody (1/50 dilution)
- single-stained with PE-Cy5 conjugated anti-CD19 antibody (1/50 dilution)
- triple-stained for CD23, CD58, and CD19 (1/50 dilution each)
- single-stained with PE conjugated isotype control antibody matched to CD23-PE (at the same concentration as anti-CD23 antibody)
- single-stained with FITC conjugated isotype control antibody matched to CD58-FITC (at the same concentration as anti-CD58 antibody)
- single-stained with PE-Cy5 conjugated isotype control antibody matched to CD19-PE-Cy5 (at the same concentration as anti-CD19 antibody)
- triple-stained with all three isotype control antibodies.
- In day three post-exposure to EBV, gently swirl flask to obtain a more uniform cell suspension. Remove 2ml from flask into a 2ml Eppendorf tube. Spin tube in a microcentrifuge at 3000 rpm at room temperature for 3 minutes.
- Aspirate supernatant and resuspend cells in FACS buffer at 5 x 106 to 1 x 107cells/ml.
- Remove eight 50μl aliquots into individual Eppendorf tubes or a 96-well V-bottom plate. Spin tubes or the plate at 1500 x g for 3 minutes.
- Discard supernatant and vortex tubes/plate. Resuspend cells in each tube/well in 50 μL of FACS buffer containing appropriate antibody dilutions prepared in 4.2.
- Incubate tubes/plate on ice in the dark for 30 minutes. Centrifuge cells at 3000 rpm for 3 minutes, remove supernatant, and add 200 μL of FACS buffer. Centrifuge cells again and remove supernatant to complete the wash. Repeat two more washes.
- Resuspend cells in 200 μL FACS buffer and acquire data using a flow cytometer, acquiring 20,000 events/tube.
- Analyze data using WinMDI (freeware for FACS data analysis on PC). Gate on live cells using forward and side scatter profiles. Then, gate live cells for expression of CD19+ (B cell marker) after comparing with cells stained with isotype control antibody matched to anti-CD19 antibody. Plot CD19+ live B cells with fluorescence intensity for CD23 on x-axis and fluorescence intensity for CD58 on y-axis. Determine CD23+ and CD58+ cells by comparing to EBV-exposed cells stained with matched isotype control antibodies. Presence of CD23hiCD58+ cells in EBV-exposed culture (Figure 2C) predicts successful outgrowth of EBV-infected growth transformed cells. In contrast, CD23hiCD58+ cells do not emerge when cells are not exposed to EBV (Figure 2A) or exposed to a non-immortalizing strain of EBV (Figure 2B). CD23hiCD58+ cells have been experimentally demonstrated to undergo proliferation and subsequently establish LCL (14).
5. Expansion and cryopreservation of LCL
- Visualization of cells by light microscopy: By a week after EBV infection, clusters of cells are visible by light microscopy. Figure 3 shows an example of early microscopic clusters (Figure 3B) in a flask.
- As time progresses, microscopic clusters become larger such that clumps are visible macroscopically in the flask. Figure 3C shows larger clusters of cells in established LCL by light microscopy.
- Feeding cells: Double the volume of culture medium in the culture flask on day 12. Subsequently, expand culture by increasing its volume 2-3 fold using complete RPMI.
- Periodicity of feeding cells should be determined based on rate of cell growth. When the culture medium turns yellow, it is generally time to replace the media as above. Typically, this occurs once per week. However, some cell lines may need to be fed more or less frequently. Expand culture to 75-100 ml over the next few weeks.
- Cryopreservation: When freezing down cells, centrifuge cells at 600 x g for 10 minutes at room temperature. Remove the supernatant and resuspend cell pellet in cold freezing medium containing 90% FBS and 10% dimethylsulfoxide at 1 x 107 cells/ml. Place tube in liquid nitrogen.
6. Representative results:
Successful outcome is predicted by the presence of CD23hiCD58+ cells. Approximately 2-3% of live cells on day 3-4 after exposure to EBV should have this profile (Figure 2C). Other sub-populations of CD23+, CD23–, CD58+, and CD58– cells observed after exposure to EBV show minimal proliferation14. Un-infected cells (Figure 2A) and cells exposed to EBV from HH514-16 cells (Figure 2B), harboring a non-immortalizing strain of EBV, do not demonstrate CD23hiCD58+cells.
You may also visualize cells by light microscopy to monitor successful outcome: As mentioned earlier, within a week after EBV infection, small clusters of cells in the flask are visible by light microscopy (Figure 3B). As time progresses and cells develop into LCL, microscopic clusters become larger (Figure 3C) such that clumps are visible macroscopically in the flask.
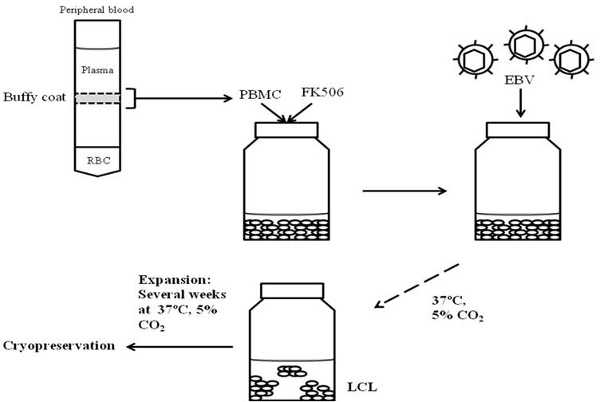
Figure 1. Workflow for generation and cryopreservation of lymphoblastoid cell lines. Peripheral blood is centrifuged through a Ficoll gradient. PBMC present in the buffy coat of an established gradient are exposed to FK506 followed by addition of EBV. EBV-exposed cells are grown at 37°C in the presence of 5% CO2 to establish and subsequently expand LCL for cryopreservation.
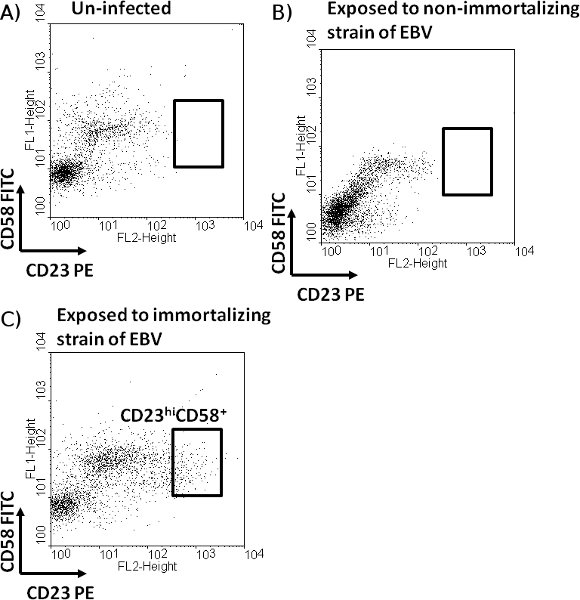
Figure 2. Identification of a sub-population of B cells expected to undergo proliferation after exposure to EBV. Un-infected PBMC (A) or PBMC exposed to EBV derived from HH514-16 cells (B) or from B95-8 cells (C) in the presence of FK506 were harvested on day 3. Cells were stained with fluorochrome-conjugated antibodies directed against CD19, CD23, and CD58. After gating on live cells, un-infected (A) and EBV-exposed (B and C) CD19+ B cells were examined for expression of CD23 and CD58. A sub-population of B cells expressing CD58 and high levels of CD23 (CD23hiCD58+), observed only in the cells exposed to transformation competent EBV (derived from B95-8 cells), is depicted.
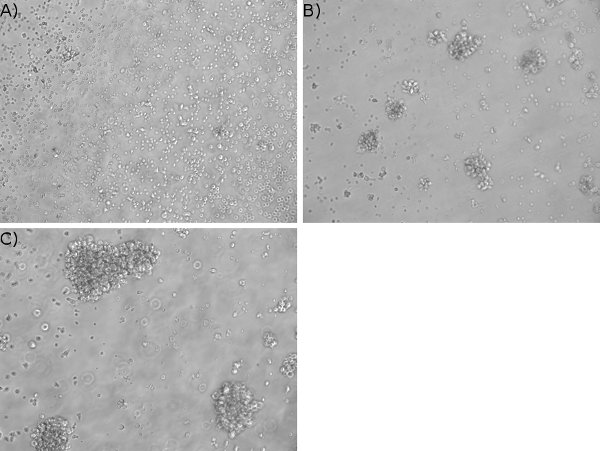
Figure 3. Visualization of cell clusters in EBV-infected cells. PBMC were treated with FK506 and infected with EBV. Un-infected (A) and EBV-exposed (B) cells were examined one week post-infection by phase contrast microscopy (10X magnification). Five-week old lymphoblastoid cell line is shown in C.
Discussion
The method described in this paper generates LCL from donor peripheral blood with rapid immortalization and cryopreservation times. Through the use of FK506, a T cell immunosuppressant, and high titers of infectious virus we are able to promote proliferation of EBV-infected B cells from peripheral blood mononuclear cells. These interventions make the described method more efficient, resulting in rapid expansion of cells for subsequent experiments.
Traditionally, growth transformation has been monitored by visualization of clusters of cells by light microscopy about a week after exposure to EBV6, 15. However, clustering of cells is not a specific indicator of EBV-mediated growth transformation. We have previously demonstrated consistent identification of the proliferating cell population via flow cytometry14, providing an accurate and specific method to determine successful outcome as early as three days after exposure of B cells to EBV.
Declarações
The authors have nothing to disclose.
Acknowledgements
This research was funded by the NIH grants K08 AI062732, K12 HD001401, and 1UL1RR024139-02 and a Child Health Research Grant from the Charles H. Hood Foundation to S.B.-M. and by the Research Foundation at the State University of New York at Stony Brook.
Materials
| Company | Catalog number | |
| Amphotericin B | Fisher Bioreagents | BP 928-250 |
| Dimethylsulfoxide | Sigma | D 2650 |
| Fetal bovine serum, heat-inactivated | Sigma | F 4135 |
| Ficoll Hypaque lymphocyte separation media | Mediatech | 25-072 |
| FK506 | A.G Scientific | F 1030 |
| IgG from mouse serum | Sigma | I 8765 |
| Mouse anti-human CD23 conjugated to PE | BD Pharmingen | 555711 |
| Mouse isotype IgG1 conjugated to PE | BD Pharmingen | 554680 |
| Mouse anti-human CD58 conjugated to FITC | Pierce Thermo Scientific | MA1-82159 |
| Mouse isotype IgG1 conjugated to FITC | BD Pharmingen | 550616 |
| Mouse anti-human CD19 conjugated to PE-Cy5 | BD Pharmingen | 555414 |
| Mouse isotype IgG1 conjugated to PE-Cy5 | Dako | X 0955 |
| Penicillin/Streptomycin | Gibco | 15140-122 |
| RPMI 1640 media | Sigma | R 8758 |
| Tetradecanoyl phorbol acetate (TPA) | Calbiochem | 407952 |
| FACS Buffer (1X Phosphate Buffered Saline + 5% Fetal Bovine Serum) |
Referências
- Thorley-Lawson, D. A., Gross, A. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 350, 1328-1337 (2004).
- Kubuschok, B. Use of spontaneous Epstein-Barr virus lymphoblastoid cell lines genetically modified to express tumor antigen as cancer vaccines: mutated p21 ras oncogene in pancreatic carcinoma as a model. Hum. Gene Ther. 13, 815-827 (2002).
- Kuppers, R. B cells under influence: transformation of B cells by Epstein-Barr virus. Nat. Rev. Immunol. 3, 801-812 (2003).
- Traggiai, E. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 10, 871-875 (2004).
- Bernasconi, N., Traggiai, E., Lanzavecchia, A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science. 298, 2199-2202 (2002).
- Oh, H. -. M. An efficient method for the rapid establishment of Epstein-Barr virus immortalization of human B lymphocytes. Cell Prolif. 36, 191-197 (2003).
- Ventura, M. Use of a simple method for the Epstein-Barr virus transformation of lymphocytes from members of large families of Reunion Island. Hum. Hered. 38, 36-43 (1988).
- Henderson, E. Efficiency of transformation of lymphocytes by Epstein-Barr virus. Virology. 76, 152-163 (1977).
- Bird, A. G. Characteristics of Epstein-Barr virus activation of human B lymphocytes. J. Exp. Med. 154, 832-839 (1981).
- Neitzel, H. A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum. Genet. 73, 320-326 (1986).
- Pelloquin, F., Lamelin, J. P., Lenoir, G. M. Human B lymphocytes immortalization by Epstein-Barr virus in the presence of cyclosporin A. In Vitro Cell Dev. Biol. 22, 689-694 (1986).
- Pressman, S., Rotter, J. I. Epstein-Barr virus transformation of cryopreserved lymphocytes: prolonged experience with technique. Am. J. Hum. Genet. 49, 467-467 (1991).
- Miller, G. Epstein-Barr virus: Transformation, cytopathic changes, and viral antigens in squirrel monkey and marmoset leukocytes. Proc. Nat. Acad. Sci. 69, 383-387 (1972).
- Megyola, C., Ye, J., Bhaduri-McIntosh, S. Identification of a sub-population of B cells that proliferates after infection with Epstein-Barr virus. Virol. J. 8, 84-84 (2011).
- Tosato, G., Cohen, J. Generation of Epstein-Barr virus (EBV)-immortalized B cell lines. Current Protocols of Immunology. Chapter 7, 22-22 (1991).

