Ex vivo Live Imaging of Single Cell Divisions in Mouse Neuroepithelium
Summary
Here we develop the tools necessary for ex vivo live imaging to trace single cell divisions in the mouse E8.5 neuroepithelium
Abstract
We developed a system that integrates live imaging of fluorescent markers and culturing slices of embryonic mouse neuroepithelium. We took advantage of existing mouse lines for genetic cell lineage tracing: a tamoxifen-inducible Cre line and a Cre reporter line expressing dsRed upon Cre-mediated recombination. By using a relatively low level of tamoxifen, we were able to induce recombination in a small number of cells, permitting us to follow individual cell divisions. Additionally, we observed the transcriptional response to Sonic Hedgehog (Shh) signaling using an Olig2-eGFP transgenic line 1-3 and we monitored formation of cilia by infecting the cultured slice with virus expressing the cilia marker, Sstr3-GFP 4. In order to image the neuroepithelium, we harvested embryos at E8.5, isolated the neural tube, mounted the neural slice in proper culturing conditions into the imaging chamber and performed time-lapse confocal imaging. Our ex vivo live imaging method enables us to trace single cell divisions to assess the relative timing of primary cilia formation and Shh response in a physiologically relevant manner. This method can be easily adapted using distinct fluorescent markers and provides the field the tools with which to monitor cell behavior in situ and in real time.
Protocol
Adult mice are euthanized by mechanical cervical dislocation. All animal procedures were approved by the IACUC and the Biosafety Committee at Emory University.
1. Embryo Generation
- Cross tamoxifen- inducible Cre line, CAGGCreER, and dsRedCre reporter line (Tg(CAG-Bgeo,-DsRed*MST)1Nagy) (Figure 1) 5,6. To monitor the relative timing of Shh response in the daughter cells, cross CAGGCreER and dsRed line with the Olig2-eGFP BAC transgenic mice (Tg(Olig2-EGFP)EK23Gsat) (Figure 1) 7,8.
- Dissolve tamoxifen in 100% ethanol, and perform intraperitoneal injection of 2.5 mg tamoxifen per 40 g of body weight into pregnant females at embryonic 6.5 (E6.5) to induce Cre expression and dsRed labeling in a small subset of cells.
- 48 hr later dissect embryos, identify dsRed positive embryos using the fluorescent microscope, transfer them to the culture dish and observe single cell divisions during ex vivo imaging (see below).
2. Whole Mouse Embryo Culture
- Dissect E8.5 embryos in pre-warmed wash medium containing DMEM/F12 (1:1) supplemented with 10% newborn calf serum and 1% Penicillin/Streptomycin (P/S) 9.
- Directly after dissection, place embryos on the 37 °C heating stage under the fluorescent microscope and identify as GFP and/or dsRed positive (see below).
- Transfer up to 2 embryos into a 500 μl drop of pre-equilibrated culture media containing 50% Rat Serum from a Sprague-Dawley male and 50% DMEM/F12 (1:1) without phenol red supplemented with L-Glutamine and 1% of 1 M HEPES in 0.85% NaCl and P/S 9.
- Apply a thin layer (0.1 cm) of equilibrated light mineral oil over the medium to prevent evaporation and transfer the culture dish containing the embryos to the 37 °C, 5% CO2 incubator.
3. Viral Infection
- In order to label cilia in the neuroepithelium, add 5-10 μl of Sstr3-GFP lentivirus, approximately 2 million virions, to a 500 μl equilibrated drop of culture medium containing an E8.5 embryo.
- After 18 hr of culture, transfer infected embryo into several drops of wash medium to wash out the virus and prepare neural tube slice.
4. Neural Tube Slice Preparation for Live Imaging
- Dissect neural tube of the E8.5 embryo in pre-warmed wash medium using a micro-knife size 0.025 mm, on a 1% agar coated dish.
- Place the isolated neural tube ventral side down in a 150 μl drop of equilibrated culture medium without phenol red on the 35 mm poly-L-lysine coated glass bottom dish.
- Put small amounts of a 1:1 mixture made from 100% pure petroleum jelly and melted candle wax (candle from IKEA) around the mounted neural tube, and gently press by a narrow piece of glass coverslip in order to immobilize the sample.
- Cover the dish with a thin layer (~0.1 cm) of equilibrated light mineral oil (Figure 2).
5. Live Imaging and Time-lapse Confocal Microscopy
- Place dish under the Nikon A1R Laser Scanning Confocal Inverted Microscope equipped with an environmental chamber that regulates temperature, set to 37 °C, and 5% CO2.
- Use 60x oil-immersion objective to record GFP labeled cilia and dsRed positive cells, while the 40x oil-immersion objective use to monitor Olig2-GFP and dsRed positive cells.
- Open the NIS Elements software to set up time lapse imaging conditions. Every 10 min acquire z-stacks of up to 25 μm with a spacing of 1.5 μm (40x objective) and up to 8 μm with a spacing of 0.4 μm (60x objective). Use 488 and 561 nm the excitation wavelengths and transmitted channel if needed. Acquire images at 512 x 512 size. Set up multiple user-defined regions of interest to perform simultaneously recording.
Optimal exposure to laser power and brightness of the image was adjusted by use low laser intensity (up to 11% for mCherry 561; up to 4.5% EGFP 405/488), scan speed 1, line average 2 and size of pin hole (61 μm for dsRED; 28.1 μm for Olig2; 72 μm for SSTR3GFP; 61.3 μm for dsRED and SSTR3GFP; 58 μm for dsRED and Olig2). The use of genetically encoded fluorescent reporters enabled us to detect the brightness with low laser power. - Analyze recorded data using Imaris 3D reconstruction software.
6. Immunofluorescence
- Fix embryos or isolated neural tubes in 4% paraformaldehyde / 0.1 M Phosphate Buffer on ice (4 °C) for 1 hr in a glass dish.
- Wash samples 2 hr in PBS on ice (4 °C) (change PBS a few times) and put in 30% sucrose / 0.1 M Phosphate Buffer over the night or until embryos sink.
- Embed in OCT, freeze on dry ice and store in -80 °C. Perform sectioning on cryostat (10 mm).
- Wash slides for 10 min in wash solution containing 1% Heat-inactivated sheep serum and 0.1% Triton X-100 in PBS.
- Dilute primary antibodies in wash solution at following concentrations: Rat monoclonal anti-RFP (5F8,) 1:200; Rabbit anti-Arl13b serum 1:1500 and mouse monoclonal anti-Arl13b 1:5 (295B/54); Rabbit anti-Olig2 1:300; Mouse monoclonals Pax6, Shh and Nkx2.2- all 1:10; and Rabbit polyclonal Ki67 1:500. Add around 150 μl per slide in flat humidified chamber, cover with parafilm to avoid drying out and leave over the night at 4 °C.
- Wash slides in wash solution three times, 20 min each time at room temperature.
- Dilute secondary antibodies Alexa Fluor 488, 568, 350 in wash solutions at 1:200 concentration. Use Hoechst 1:3,000 or TO-PRO-3 1:500 to stain nuclei. Add 150 μl per slide, leave 1 hr at room temperature in humidified chamber protected from light.
- Wash slides in wash solution two times, 30 min each time at room temperature.
- Mount slides in 80% ProLong Gold anti-fade reagent and view within 24 hr.
Representative Results
Here we performed ex vivo live imaging of single cell divisions within the E8.5 mouse neuroepithelium. To label individual cells, we induced Cre recombinase in a subset of cells containing a Cre reporter line that expressed dsRed upon recombination 5,6(Figure 3A). Thus, 48 hr later we were able to observe single cell divisions during ex vivo imaging (Figures 4A-D). Concomitantly, we monitored when the labeled cells became Shh-responsive by including a Shh transcriptional reporter modified BAC transgenic Olig2-eGFP line (Figures 3C-D; Figures 4G-N) 1-3. To observe cilia formation we generated Sstr3-GFP lentivirus to infect embryos in culture (Figure 3B; Figures 4A-D) 4. Immunofluorescence was carried out to confirm in vivo results (Figures 4E-F).
The time-lapse confocal imaging observation of the dsRedCre reporter embryos expressing Olig2-eGFP or infected with Sstr3-GFP lentivirus during in vitro culture are showed in Figure 5 10. In order to be sure that the Sstr3-GFP expression we were using to visualize cilia was not interfering with any underlying biological process, we corroborated our live imaging data with fixed wild-type embryo sections. Because we found the same percentage of asynchrony in cilia formation and Shh response, it indicates the SSTR3-GFP virus did not impact these processes (Figure 5A).
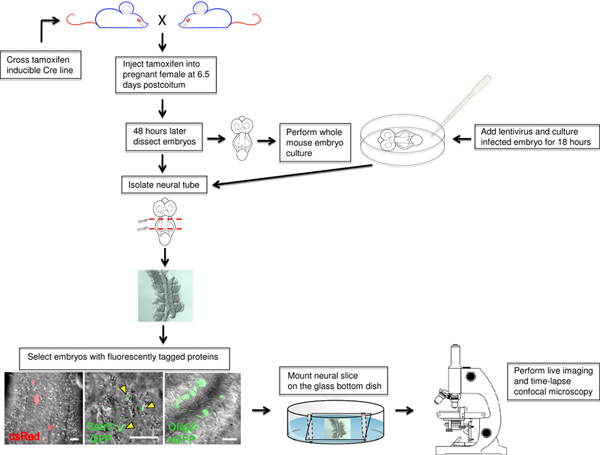
Figure 1. Flow chart of the ex vivo live imaging procedure using mouse neuroepithelium. The final mouse cross and tamoxifen treatment are depicted followed by the whole mouse embryo culture method. Embryos expressing fluorescently tagged proteins are selected and prepared for live imaging. Click here to view larger figure.
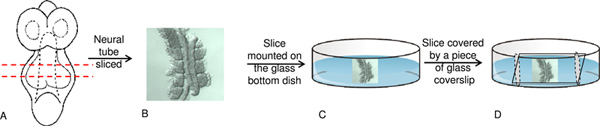
Figure 2. Neural tube slice preparation for live imaging. A) E8.5 unturned embryo with first appearance of somite pairs. B) Neural tube dissected in pre-warmed medium using a micro-knife. C) Neural tube slice is mounted ventral side down on the poly-L-lysine coated glass bottom dish. D) Small amount of a mixture made from petroleum jelly and wax applied around the neural tube and gently covered by a narrow piece of glass coverslip. Click here to view larger figure.
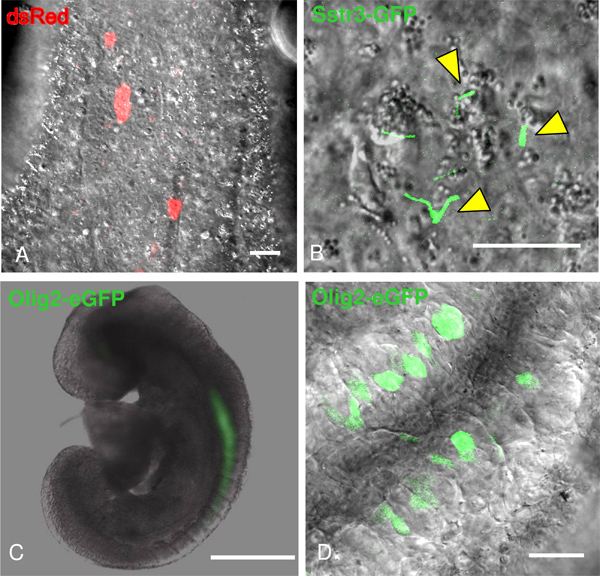
Figure 3. Visualizing fluorescently tagged proteins in live cells of the neural tube with confocal microscopy. A) DsRed labels individual cells. B) SSTR3-GFP lentivirus expressed in primary cilia as they form (yellow arrowheads). C-D) E8.5 embryo carrying Olig2-GFP shows GFP expression within neuroepithelium. Bar is 30 μm (A, D), 15 μm (B) and 1.5 μm (C).
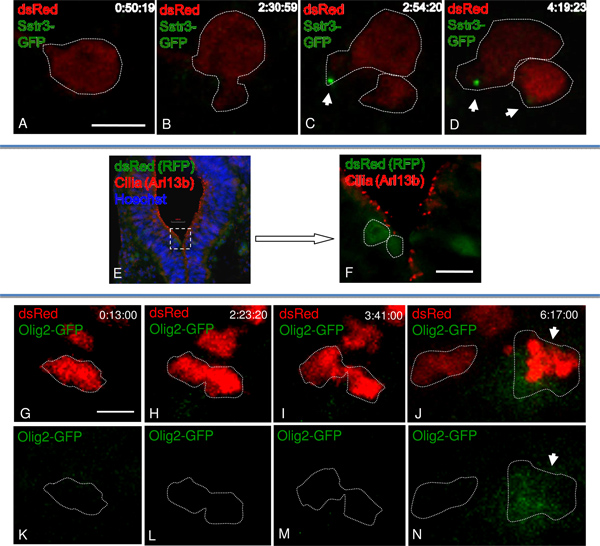
Figure 4. Monitoring cilia formation and Shh signaling in dividing cells of the developing neural tube. A-D) Single dsRed positive cell undergoing division shows a cilium forms in one daughter cell prior to the other cell (C, white arrow). The movie was imaged at a rate of one frame every 10 min. E-F) Immunofluorescence using antibodies against RFP in green (for dsRed lineage tracing) and Arl13b in red (for cilia). Enlargement of boxed area (E), without Hoechst (F). G-N) The dsRed positive cell undergoes division (I-M). The Olig2-GFP is expressed only in one daughter (J, N). The recording was imaged at a rate of one frame every 10 min. Bar is 5 μm (A-D), 50 μm (F), 25 μm (G-N).
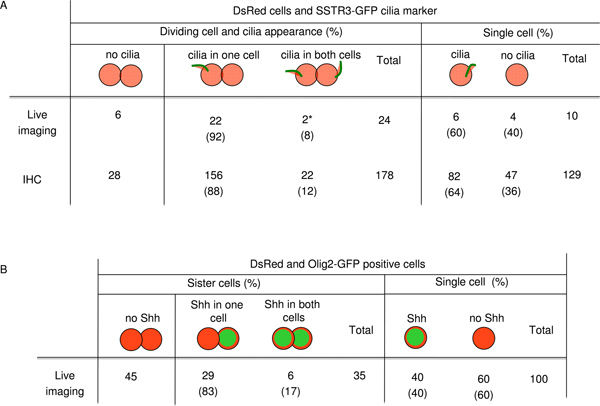
Figure 5. Counting of dsRed positive cell with cilia and Shh appearance. A) Number of dsRed cells undergoing division and cilia localization. Symbol * means by live imaging cilia formation was synchronous. B) Number of dsRed and Olig2-GFP positive cells undergoing division and Shh signaling.
Discussion
Our ex vivo system enables us to directly observe single cell divisions within the developing neuroepithelium in real time. As an example we examined cell divisions within the mouse embryonic neural tube and monitored either cilium formation or Shh response. We confirmed our imaging results (n = 24) were consistent with results from fixed sections (n = 178) indicating our technique provides physiologically relevant data.
Our technique relies on Cre induction being limited to a subset of cells so the dose of tamoxifen must be precise. We worked out the dosing for labeling single cells. We believe it would be easy to label small clones of cells with a slight increase in the dose based on the initial analysis of the CAGGcreER line, which showed a dose dependent response to tamoxifen 6. Additionally, the embryo culture conditions are critical, as abnormal development occurs if the conditions are off. We confirmed that development proceeded normally under the culture conditions we used, which are known to work for at least 36 hr 11.
The inducible Cre and Cre reporter lines we use are publically available so can be easily obtained making this technique quite adaptable to researchers with a variety of questions. The real power of our technique is that in genetically labeling single cells, our approach can be used to interrogate single cell behavior in the many existing and future mutant mouse lines. This will be critical in understanding what is truly going wrong in many of these lines as direct observation of cells within the mammalian embryo during development has not been extensively examined. Our incorporaton of genetically encoded fluorescent reporters to detect brightness with low laser power provides a real advantage for such observation. It is easy to imagine fluorescently tagged proteins marking specific cellular organelles or other transcriptional reporters being integrated in the future.
We were interested in the relative timing of primary cilium formation and Shh response in the daughter cells of a dividing cell. However, the transgenic line that enabled us to monitor Shh response expressed Olig2-eGFP throughout the cytoplasm, precluding us from seeing the marker of the primary cilium, Sstr3-GFP in the same cell. Thus, we would have greatly benefited from either a nuclear restricted Olig2-eGFP or a spectrally distinct Sstr3 fluorescent protein fusion.
In addition to using targeted and transgenic line, we showed that our technique can be adapted using viral constructs and that infecting the neuroepithelium in culture provides useful data. This will be useful to investigators in providing a quicker method than generation of a genetically modified mouse line. Additionally, the approach can validate the generation of such a line; indeed, our data argue a transgenic line expressing Sstr3-GFP will be of great use to the field.
Our method provides tools with which the field can more immediately monitor cell behavior. Coupled to the rich resources of mouse mutants we expect this method to be especially powerful in examining an array of cellular behaviors in situ.
Declarações
The authors have nothing to disclose.
Acknowledgements
This research project was supported by an ARRA Supplement, 5 R01 NS056380. Additional support was provided through the Viral Vector Core and the Microscopy Core of the Emory Neuroscience NINDS Core Facilities grant, P30NS055077. We thank the Emory Transgenic Mouse and Gene Targeting Core for deriving the mouse line from GENSAT; Greg Pazour for the stable SSTR3-GFP IMCD3 cell line; and Bradley Yoder for the Sstr3-GFP lentiviral construct. Monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD, and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. All animal procedures were approved by the IACUC and the Biosafety Committee at Emory University.
Materials
| Name of the reagent | Company | Catalogue number | Comments (optional) |
| Z/RED line (STOCK Tg(CAG-Bgeo,-DsRed*MST)1Nagy/J | Jackson Laboratory | 005438 | |
| Olig2-eGFP line (STOCK Tg(Olig2-EGFP)EK23Gsat/Mmcd | MMRRC, | 010555-UCD | |
| CAGGCreER | Jackson Laboratory | 003724 | |
| Tamoxifen | Sigma | T5648 | |
| DMEM/F12 (1:1) | GIBCO | 21041-025 | |
| Newborn calf serum | Lonza | 14-416F | |
| Penicillin/Streptomycin | Invitrogen | 15140-122 | |
| Rat Serum SD male | Harlan Bioproducts | 4520 | |
| 1M Hepes | BioWhittaker | 17-737E | |
| L-Glutamine | GIBCO | 21041-025 | |
| Light mineral oil | Sigma | M8410 | |
| Sstr3-GFP lentivirus | Emory Viral Core | ||
| Micro-knife, size 0.025 mm | Electron Microscopy Sciences | 62091 | |
| 35 mm poly-L-lysine coated glass bottom dish | MatTek | P35GC-0-10-C | |
| 100% petroleum jelly | Kroger | FL9958c | |
| A1R Laser Scanning Confocal Inverted Microscope | Nikon | ||
| NIS Elements software | Nikon | ||
| Imaris 3D software | Bitplane AG | Imaris 7.2.3 | |
| OCT | Tissue-Tek | 4583 | |
| Cryostat | Leica | CM1850 | |
| Heat-inactivated sheep serum | Invitrogen | 16210-072 | |
| Triton X-100 | Fisher Scientific | BP151 | |
| Parafolmaldehyde | Sigma | P6148 | |
| Phosphate Buffer | Lab made | ||
| Rat monoclonal anti-RFP (5F8) | Chromotek | 110411 | |
| Rabbit anti-Arl13b serum | NeuroMab | ||
| mouse monoclonal anti-Arl13b 1:5 | NeuroMab | ||
| Rabbit anti-Olig2 | Chemicon | AB9610 | |
| Mouse monoclonals Pax6 | Developmental Hybridoma Bank | Pax6 | |
| Mouse monoclonalsShh | Developmental Hybridoma Bank | 5E1 | |
| Mouse monoclonals Nkx2.2 | Developmental Hybridoma Bank | 74.5A5 | |
| Rabbit polyclonal Ki67 | Abcam | AB15580 | |
| Alexa Fluor 488 | Molecular Probes | A11029 | |
| Alexa Fluor 568 | Molecular Probes | A11031 | |
| Alexa Fluor 350 | Molecular Probes | A11046 | |
| Hoechst | Fisher | AC22989 | |
| TO-PRO-3 | Invitrogen | T3605 | |
| ProLong Gold anti-fade reagent | Invitrogen | P36934 |
Referências
- Yamada, T., Pfaff, S. L., Edlund, T., Jessell, T. M. Control of cell pattern in the neural tube: motor neuron induction by diffusible factors from notochord and floor plate. Cell. 73, 673-686 (1993).
- Ericson, J., Muhr, J., Placzek, M., Lints, T., Jessell, T. M., Edlund, T. Sonic hedgehog induces the differentiation of ventral forebrain neurons: A common signal for ventral patterning within the neural tube. Cell. 81, 747-756 (1995).
- Briscoe, J. A. Homeodomain Protein Code Specifies Progenitor Cell Identity and Neuronal Fate in the Ventral Neural Tube. Cell. 101, 435-445 (2000).
- Berbari, N. F., Johnson, A. D., Lewis, J. S., Askwith, C. C., Mykytyn, K. Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol. Biol. Cell. 19 (4), 1540-1547 (2008).
- Vintersten, K. Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis. 40 (4), 241-246 (2004).
- Hayashi, S., McMahon, A. P. Efficient recombination in diverse tissue by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 244 (2), 305-318 (2002).
- Gong, S., Zheng, C., Doughty, M. L., Losos, K., Didkovsky, N., Schambra, U. B., Nowak, N. J., Joyner, A., Leblanc, G., Hatten, M. E., Heintz, N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 425 (6961), 917-925 (2003).
- Mukouyama, Y. S., Deneen, B., Lukaszewicz, A., Novitch, B. G., Wichterle, H., Jessell, T. M., Anderson, D. J. Olig2+ neuroepithelial motoneuron progenitors are not multipotent stem cells in vivo. Proc. Natl. Acad. Sci. U.S.A. 103 (5), 1551-1556 (2006).
- Jones, E. A. V., Crotty, D., Kulesa, P. M., Waters, C. W., Baron, M. H., Fraser, S. E., Dickinson, M. E. Dynamic in vivo imaging of postimplantation mammalian embryos using whole embryo culture. Genesis. 34, 228-235 (2002).
- Piotrowska-Nitsche, K., Caspary, T. Live imaging of individual cell divisions in mouse neuroepithelium shows asymmetry in cilium formation and Sonic hedgehog response. Cilia. 1 (6), (2012).
- Nowotschin, S., Ferrer-Vaquer, A., Hadjantonakis, A. -. K. Imaging mouse development with confocal time-lapse microscopy. Methods in Enzymology. 476, 351-377 (2010).

