Visualization of Twitching Motility and Characterization of the Role of the PilG de Xylella fastidiosa
Summary
In this study, a nano-microfluidic flow chamber was employed to visualize and functionally characterize the twitching motility of Xylella fastidiosa, a bacterium that causes Pierce's disease in grapevine.
Abstract
Xylella fastidiosa is a Gram-negative non-flagellated bacterium that causes a number of economically important diseases of plants. The twitching motility provides X. fastidiosa a means for long-distance intra-plant movement and colonization, contributing toward pathogenicity in X. fastidiosa. The twitching motility of X. fastidiosa is operated by type IV pili. Type IV pili of Xylella fastidiosa are regulated by pilG, a chemotaxis regulator in Pil-Chp operon encoding proteins that are involved with signal transduction pathways. To elucidate the roles of pilG in the twitching motility of X. fastidiosa, a pilG-deficient mutant XfΔpilG and its complementary strain XfΔpilG-C containing native pilG were developed. A microfluidic chambers integrated with a time-lapse image recording system was used to observe twitching motility in XfΔpilG, XfΔpilG-C and its wild type strain. Using this recording system, it permits long-term spatial and temporal observations of aggregation, migration of individual cells and populations of bacteria via twitching motility. X. fastidiosa wild type and complementary XfΔpilG-C strain showed typical twitching motility characteristics directly observed in the microfluidic flow chambers, whereas mutant XfΔpliG exhibited the twitching deficient phenotype. This study demonstrates that pilG contributes to the twitching motility of X. fastidiosa. The microfluidic flow chamber is used as a means for observing twitching motility.
Introduction
Xylella fastidiosa is a Gram-negative non-flagellated, pathogenic bacterium that causes a number of economically important crop diseases, including Pierce's disease in grapevine (Vitis vinifera L.)1,2, 3. This bacterium is limited to the water-conducting xylem vessels. Infection of grapevine causes the blockage of xylem vessels and results in water stress and nutritional deficiencies3. Successful colonization depends on the ability of the bacterium to move from the initial site of infection to the rest of the plant3. Twitching motility is a means of flagellar-independent bacterial movement through the extension, attachment, and retraction of the polar type IV pili4 that has been characterized in X. fastidiosa5,6,7.
The twitching motility has been observed by laser tweezers and atomic force microscopy (AFM) 8,9,10. Using these techniques, twitching motilities generated by type IV pilus of N. gonorrhoeae and P. aeruginosa were characterized by fluorescently labeling pili and capturing their movements microscopically. Although both methods have detailed the adhesive force of individual bacteria, the procedures are complicated and time consuming9,10. The microfluidic chambers were used to observe long-distance migration of individual cells as well as small aggregates of bacterial cells5,6. These chambers were designed as a microfabricated-nano-channel in a plate integrated with a time-lapse image recording system11,12,13,14. Microfluidic chamber devices offer several advantages for studying the movement behavior and cell-cell interactions of bacteria: (i) it provides an integrated platform with multiple channel capabilities; (ii) it can examine the motions and aggregations of single cells in the nano-scale features of bacteria; (iii) it allows for direct microscopic image recording of bacterial cells and time-lapse analysis, (iv) it provides long-term spatial and temporal observations of individual and/or populations of bacteria in a micro-environment; (v) the flow rate of culture medium in a channel can be precisely controlled and (vi) only a very small volume (1 ml) of culture medium is required for each experiment.
Recently, the microfluidic flow system has been employed to investigate the behaviors of bacterial cells under various microenvironments 14,15,16. The adhesiveness and the surface attachment of E. coli15, X. fastidiosa16, and Acidovorax citrulli14 to glass surfaces were assessed using microfluidic chambers. The aggregation and biofilm formation mediated by type IV pili of Acidovorax citrulli were analyzed14. Furthermore, the motion of A. citrulli observed under flow conditions demonstrated that the type IV pili may play important roles in the colonization and spread of A. citrulli in xylem vessels under sap flow conditions. The twitching motilities of Pseudomonas aeruginosa and X. fastidiosa cells were successfully observed against a fluid current in a microfabricated flow chamber5,6,17. Type IV pilus deficient pilB and pilQ mutants of X. fastidiosa were found to profoundly alter the speed of twitching motility under the flow conditions in microfluidic devices5,6,18. The studies conducted on bacterial adhesion and motility in microfluidic devices indicated that the microfluidic chambers are particularly suitable for analyzing the twitching motility and migration of pili-mediated bacteria in vitro. These results explain the twitching-mediated migration mechanism which facilitates cell-cell attachment, aggregation and colonization within the host, eventually lead to systemic infection.
Pil-Chp operon of X. fastidiosa contains pilG, pilI, pilJ, pilL, chpB and chpC which encode signal transduction pathways20. The transmembrane chemoreceptors bind chemical stimuli in the periplasmic domain and activate a signaling cascade in their cytoplasmic portion to ultimately control bacterial twitching motility. In the Pil-Chp operon of X. fastidiosa, a phospho-shuttle protein PilG is a homologue to CheY. In E. coli and P. aeruginosa, CheY is the response regulator in chemotaxis systems that interact with the flagella motor proteins19,21. Although the contributions of the Pil-Chp operon toward virulence in X. fastidiosa were examined recently20, the role of pilG in chemotaxis operon in response to the environmental signals and to regulated/motor type IV pili of X. fastidiosa is unclear. To elucidate the insight of chemotaxis regulator pilG in the activity of twitching motility of X. fastidiosa, a microfluidic chamber is used to assess the twitching motility of X. fastidiosa. The pilG of X. fastidiosa is characterizedby comparing the phenotypes of a deletion mutant XfΔpliG, complementary strain XfΔpliG -C and its wild type in vitro. The results highlight the role of pilG in the twitching motility of X. fastidiosa.
Protocol
1. The Peripheral Fringe of Bacterial Colony
- Grow X. fastidiosa (Xf) Temecula wild type22, pilG deletion mutant XfΔpliG (using previously described deletion strategy23), and its complementary XfΔpliG-C (using previously described chromosome-based genetic complementation strategy24) on PD2 medium agar plates25 at 28 °C for 5-7 days.
- Autoclave cellophane (1 x 1 cm2) in water at 121 °C (249 °F) for 15 min. Pick up one piece of cellophane, drain the water by touching one corner of cellophane on an empty Petri dish, carefully lay the cellophane over 15% of the agar surface and air dry.
- Pick up individual X. fastidiosa colonies with sterile rounded toothpicks and spot cells aseptically onto a sterilized sheet of cellophane overlaid on the 15% of agar surface in the agar plates. Incubate the plates at 28 °C for 2-3 days.
- Examine the edge morphology of the colonies using a dissecting microscope with a 2X objective lens and a 10X ocular lens. Photograph the peripheral fringe around colonies.
2. Microscopy and Microfluidic Flow Chambers
- Fabricate microfluidic devices using photo-lithographic procedures similar to those previously described5,18. Design four parallel channels with computer-assisted design software on master silicon wafer using standard lithographic methods26.
- Create the microfluidic chambers from silicon wafer master with polydimethylsiloxane (PDMS). Pour unpolymerized PDMS over the silicon wafer master and cure it at 60 °C for 1 hr. Peel off the PDMS replica from the wafer master and trim the PDMS replica with a blade into a 22 mm x 40 mm as the same size of a glass coverslip.
- Expose the PDMS replica, a glass coverslip (22 x 40 mm2), and a microscope slide (51 x 76 mm2) to air plasma at 30 W for 2 min27. Sandwich the PDMS body between the glass coverslip and the glass microscope to build the microfluidic chamber.
- Drill a hole (5.5 mm diameter) through the PDMS at each end of the patterned-channel. Cut the silicone rubber tubing into 12-20 cm long. Insert one end of the silicone rubber tubing (5.1 mm outside diameter, 2.1 mm inside diameter, 0.8 mm wall) into each opening end of the channels of the PDMS replica, and seal it with unpolymerized PDMS at 60 °C for 1 hr.
- Connect another end of the tubing to the barbed end of plastic luer connectors. Wrap the assembled microfluidic chambers with the aluminum foil and autoclave them for 20 min.
- Collect bacteria cells of X. fastidiosa wild type, mutant XfΔpliG, and complementary XfΔpliG-C via scraping, using disposable inoculating loops from PD2 medium plates. Adjust cell density to an optical density of 0.05 at 600 nm in PD2 broth as described previously23. Collect the bacterial cell solution into a 1 ml Gastight syringe.
- Mount the microfluidic device on an inverted microscope stage. Connect an inlet tube to the 5 ml Gastight syringe containing PD2 broth. Fit the 5 ml Gastight syringe with the syringe pumps.
- Connect the outlet tube to a waste reservoir. Maintain a medium flow rate of 0.2 µl min-1 for 30 min to stabilize the system.
- Connect side-inlet tubes to a 1 ml Gastight syringe containing the bacterial cell solution. Flush the bacterial cell solution through the rubber tubing until the channel is reached. Maintain a medium flow rate of 0.2 µl min-1 for another 30 min in order to flush unbound cells from the chamber prior to image capture.
- Mount the microscope shutter under the field-adjusted part of the microscope to control the light. Connect the shutter to the shutter control system and connect the shutter control system to the computer.
- Mount a digital camera to the video port of the microscope and connect it to the computer. Run the time-lapse recording software, select the "shutter" function from the menu, and recognize automatically the installed shutter as default in the software to establish connections to the shutter with the software.
- Select the "digital camera" function from the menu of the time-lapse recording software to automatically recognize the digital camera as the default capture device in the software and establish communication with the digital camera with the software.
- Locate the bacterial cells in one of the channels using 20X phase-contrast optics, then switch to the 40X objective lens prior to image capture.
- Run the time-lapse recording software, select the "image acquisition" function using default parameters from the menu to acquire the images from the microscope. Next, open the "Acquire time-lapse" function and set the time interval to 30 sec5,18,28 for duration of 6-24 hr depending on experiment needed to observe twitching motility of X. fastidiosa5,18,28. Click "OK" to start the time-lapse recording. Click "Stack function" from menu, select "save as" to stack the images in the destination folder on the computer after finishing the recording.
- For multiple channels, capture time-lapse images from the first channel every 30 sec for 6 hr. Move the objective lens of the microscope to the next channel to locate the target cells. Repeat the time-lapse function as described above to capture images in each of four channels sequentially if experiment is set up to utilize four channels. Continue the time-lapse image capture for as long as three consecutive days. All experiments were conducted at room temperature (23 ± 2 °C).
- Compile the time-lapse images into a video file using the time-lapse recording imaging software. Run time-lapse recording software, click "Stack function" from the menu, and select "open stack function" to open the stacked files from the computer.
- Start the "Make movie" function from the stack module, selecting all images and choosing the "AVI" output format. Click "save as" to store the video file in the destination folder on the computer.
- Select the compiled movies from the destination folder on the computer and play them. Then, observe the motility of the single cells through the resulting visualization of the twitching motility activity of bacteria cells in the generated video files.
Representative Results
The presence of a peripheral colony fringe indicative of type IV pilus-mediated twitching motility, was observed in the colonies of X. fastidiosa wild type and complementary XfΔpliG-C strain (Figure 1). Mutant XfΔpliG, however, did not exhibit a fringe around the periphery of the colonies (Figure 1). Time-lapse imaging of bacterial cells in nano-microfluidic flow chambers revealed that twitching motility was observed in both wild type X. fastidiosa and the complementary XfΔpliG-C (Supplemental V1, V3), whereas XfΔpilG mutant cells did not exhibit twitching motility throughout the experiment (Supplemental V2). Cells of mutant XfΔpilG formed relatively small loose aggregates in PD2 broth (Supplemental V2). In contrast, cells of X. fastidiosa wild type and complementary XfΔpilG-C developed larger aggregates in PD2 broth (Figure 2, (Supplemental V1, V3).
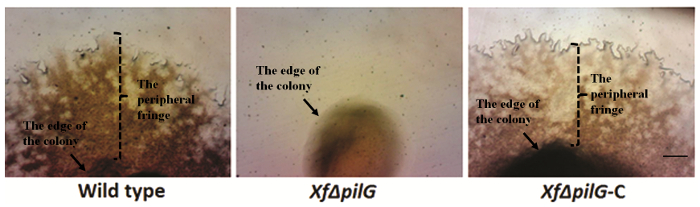
Figure 1: The peripheral fringe of bacterial colony. Colony margin characteristics of X. fastidiosa from wild type, mutant XfΔpilG, and complementary XfΔpilG-C grown on PD2 agar covered with a sterilized sheet of cellophane. With the exception of mutant XfΔpilG, all colonies exhibited a peripheral fringe, indicating type IV pilus-mediated twitching motility. Pictures were taken after 5 days of growth on culture medium. Magnification bar, 0.5 mm. Please click here to view a larger version of this figure.
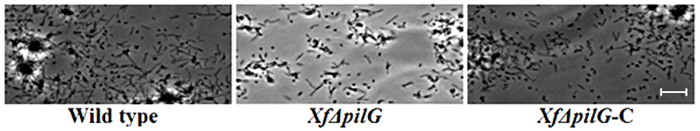
Figure 2: The twitching motility of X. fastidiosa cells in nano-microfluidic flow chamber. The twitching motility of all tested strain cells was recorded during 6 days of observation. The assessments were conducted from three independent video segments. Magnification bar, 20 µm.
Note: The twitching motility of X. fastidiosa cells is characterized by single cell movement across glass surfaces through the extension, attachment, and retraction of the polar type IV pili. The single cell was observed in the migration preferentially against a fluid current in a microfabricated flow chamber. Please click here to view a larger version of this figure.

Supplemental Figure 1: A four-channel microfluidic flow chamber. Each channel with media in and media out connectors at each end. Please click here to view a larger version of this figure.
Supplemental Movie 1: Twitching motility. (Right click to download). Twitching of wild-type X. fastidiosa in a microfluidic flow chamber.
Supplemental Movie 2: Impaired twitching motility. (Right click to download). Motility of Xf mutant. XfΔpilG observed in amicrofluidic flow chamber.
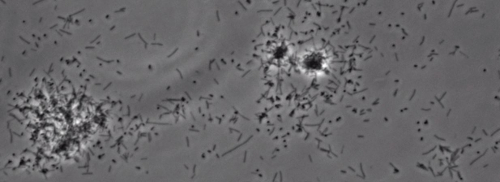
Supplemental Movie 3: Restored twitching motility. (Right click to download). Twitching motility of Xf complementary strain. XfΔpilG-C observed in a microfluidic flow chamber.
Discussion
In this study, we characterized the motion behavior of X. fastidiosa PilG mutant XfΔpilG and its complementary XfΔpilG-C strains in newly designed multiple parallel-nano-channel microfluidic chambers. The newly designed microfluidic chambers can have up to four parallel chambers with 100 µm nano-channel in width compared to earlier designs with only a single 50 µm wide channel18. The improved wider nano-channel facilitates the introduction of bacterial cells with the flowing of the media. In addition, this microfluidics chamber is 1) simple to build and assemble; 2) relatively inexpensive; and 3) easily applicable to varying experimental requirements. As a result, this chamber design permits the long-term observation of the motions of the bacterial cells under varieties experimental microenvironment.
Stabilizing the flow of the currents through the microfluidic channel is the critical step to creating an intact flowing-microfluidic microenvironment for the observation of the motility of the bacterial cells under a variety of experimental environments. The flushing of microfluidic chambers and connecting tubing with the media prior to the introduction of bacterial cells is also an important step to stabilize the flow in the chamber. However, the high speed of the flow will flush the bacterial cells out of the chamber without retaining cells in the course of the chamber. The proper speed of the media flowing through the microfluidic channels needs to be adjusted using a syringe pump. During the experiments in this study, the flowing was set and stabilized by the pump at 0.2 to 1 µl min-1 for at least 30 min prior to introducing the bacterial cells into the channel. Once the cells were introduced into the microfluidic channels, the medium flow was maintained at 0.2 µl min-1 for 30 to 60 min to stabilize the system and remove nonattached cells. It is very important to stabilize the flow and to keep the background clear in order to observe the motion of the bacterial cells. Images were captured every 30 sec to confirm the twitching motility activity of the introduced cells maintaining a constant flow speed at 0.2 µl min-1. If the experiments require the collection of the bacterial cells in the chamber, the cells can be flushed out by gradually increasing the flow rates of media from 0.2 to 110 µl min-1 by adjusting the syringe pump speed.
The bacterial cell activities in the microfluidic chambers were assessed through the inverted microscope using 40X phase-contrast optics and time-lapse images recorded with a digital camera, which was controlled by the imaging software. The flow speed and the interval time for image capture can be adjusted accordingly with experimental requirements. However, in most cases, flow speed for a medium flow rate is set to 0.2 µl min-1 with time-lapse images recorded every 30 sec for the pilus-mediated bacterial cells. In other instances, if the medium flow rate is increased to 0.1 µl min-1 during the course of experimentation, the behavior of the bacterial cells will be recorded by capturing images every 10 to 15 sec accordingly. The time-lapse images were taken every 30 sec for duration of 6-24 hr and saved as a source images files for each tested strain. The videos were then complied from the images taken from 6-8 hr from each strain, which shown contrast phenotypes in twitching motilities between mutant and wild type/complement strains (Supplemental V1, V2, V3).
The significance of the nano-microfluidic chamber devices over macroscale parallel-plate flow chambers includes the direct examination of the motions and the aggregations of single cells of bacteria. In addition to low-cost5 and low reagent and sample volume requirements, the advantages of microfluidic chambers are the ease of construction of a flow microenvironment for bacterial culture and accurate control over fluid flow rate. The multiple parallel channels permit the observation of differential bacterial strains in a single experiment setup which provides compatible data for analysis. The flagellar-independent twitching motility of the bacteria with the polar pili is particularly suitable for analysis in this nano-microfluidic chamber. However, this microfluidic chamber is less suitable for flagellar-dependent bacterial movement, in which the movement of bacteria is usually too quick and exhibits random directions. This limitation sometimes could be compromised by adjusting the medium flow rate to 0.05 µl min-1 and changing the time-lapse image capture rate at every 1 to 2 sec for 1 hr to analyze the motions of the flagellar-mediated bacteria in the nano-scale environment.
Microfluidic chamber devices used herein provide direct visual evidence for functional evaluation of the PilG responsible for motion behavior in vitro. Additionally, this study also reveals that cell to cell aggregation through the twitching motility is essential for biofilm formation, signaling and pathogenicity in X. fastidiosa. Visualization of bacteria twitching motility by microfluidic devices provides a new approach to study gene function involving behavior in bacteria which is not easily measured by other assay methods. This approach can be applied to other bacterial systems. The microfluidic chamber devices provide a flow system for characterizing the physiological behavior of bacterial cells associated with cell-cell attachments, cell aggregations, motion patterns and biofilm formation.
Declarações
The authors have nothing to disclose.
Acknowledgements
This study was supported by United States Department of Agriculture, Agricultural Research Service. Trade names or commercial products in this publication are mentioned solely for the purpose of providing specific information and does not imply recommendation or endorsement by the United States Department of Agriculture. USDA is an equal opportunity provider and employer.
Materials
| Biology materials | |||
| X. fastidiosa (Xf) Temecula wild type | Costa, H. S., et al., 2004 22 | ||
| pilG deletion mutant XfΔpliG | Shi, X. Y., et al., 2007 26 | ||
| pilG complementary strain XfΔpliG-C | Davis, M. J., wt al. 1998 23 | ||
| Physical materials and equipment | |||
| Disposable inoculating loops | VWR international, Radnor, PA | #22-363-607 | quantitative procedures such as bacterial collection |
| Polydimethylsiloxane (PDMS) | Dow Corning Corporation | #0002709226 | Sylgard 184 silicone Elastomeric Kits |
| AmScope MD2000 digital camera | AmScope, Irvine, CA | SE305R-AZ-E | Image, video recording and measurement |
| Tubes line | Edgewood, NY | #T4300 | Connected to the syringe and microfluidic chamber |
| Plastic luer connectors | Edgewood, NY | Connected to the syringe and microfluidic chamber | |
| Syringe pumps | Pico Plus, Harvard Apparatus, MA | #702209 | The flow rate can be adjusted while the pump is running. |
| Syringes | Gastight, Hemilton Company, Reno, NV | #1005 | Provide the flowing broth |
| Inverted Olympus IMT-2 microscope | Olympus | IMT-2 FLuoro PHase | Image observation and recording |
| SPOT-RT digital camera | Diagnostic Instruments, Inc., MI | RT230 | Image, video recording and measurement |
| Microscope Shutter | The UNIBLITZ, US | #LS2T2 | Control camera’s exposure time |
| Microscope Shutter Control system | The UNIBLITZ, US | VCM-D1 | VCM-D1 Single Channel CE/UL/CSA Approved Shutter Driver |
| MetaMorph Image software | Universal Imaging Corp., PA | Real-time super-resolution image processing |
Referências
- Purcell, A. H., Hopkins, D. L. Fastidious xylem-limited bacterial plant pathogens. Annu. Rev. Phytopathol. 34, 131-151 (1996).
- Purcell, A. H. Xylella fastidiosa, a regional problem or global threat. J. Plant Pathology. 79, 99-105 (1997).
- Hopkins, D. L. Xylella fastidiosa: Xylem-limited bacterial pathogen of plants. Annu. Rev. Phytopathol. 27, 271-290 (1989).
- Mattick, J. S. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289-314 (2002).
- Meng, Y., et al. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J. Bacteriol. 187, 5560-5567 (2005).
- Li, Y., et al. Type I and type IV pili of Xylella fastidiosa affect twitching motility, biofilm formation and cell-cell aggregation. Microbiology. 153, 719-726 (2007).
- Simpson, A. J. G., et al. The genome sequence of the plant pathogen Xylella fastidiosa. Nature. 406, 151-157 (2000).
- Maier, B., Potter, L., So, M., Long, C. D., Seifert, H. S., Sheetz, M. P. Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. USA. 99, 16012-16017 (2002).
- Touhami, A., Jericho, M. H., Boyd, J. M., Beveridge, T. J. Nanoscale characterization and determination of adhesion forces of Pseudomonas aeruginosa pili by using atomic force microscopy. J. Bacteriol. 188, 370-377 (2006).
- Skerker, J. M., Berg, H. C. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA. 98, 6901-6904 (2001).
- Brown, D. C., Larson, R. S. Improvements to parallel plate flow chambers to reduce reagent and cellular requirements. BMC Immunol. 2, 9 (2001).
- Thomas, W. E., Nilsson, L. M., Forero, M., Sokurenko, E. V., Vogel, V. Shear-dependent ‘stick-and-roll’ adhesion of type 1 fimbriated Escherichia coli. Mol. Microbiol. 53, 1545-1557 (2004).
- Thomas, W. E., Trintchina, E., Forero, M., Vogel, V., Sokurenko, E. V. Bacterial adhesion to target cells enhanced by shear force. Cell. 109, 913-923 (2002).
- Bahar, O., Fuente, D. L., Burdman, S. Assessing adhesion, biofilm formation and motility of Acidovorax citrulli using microfluidic flow chambers. FEMS Microbiol. Lett. 312, 33-39 (2010).
- Thomas, W. E. Using a laminar flow system to explain shear-enhanced bacterial adhesion. Proceedings of ICMM2005, Third International Conference on Microchannels and Mini-channels. , 751-759 (2005).
- Fuente, D. L., et al. Assessing adhesion forces of type I and type IV pili of Xylella fastidiosa bacteria by use of a microfluidic flow chamber. Appl. Environ. Microbiol. 73, 2690-2696 (2007).
- DeLange, P. A., Collins, T. L., Pierce, G. E., Robinson, J. B. PilJ localizes to cell poles and is required for type IV pilus extension in Pseudomonas aeruginosa. Curr Microbiol. 55, 389-395 (2007).
- Fuente, D. L., Burr, T. J., Hoch, H. C. Mutations in type I and type IV pilus biosynthetic genes affect twitching motility rates in Xylella fastidiosa. J. Bacteriol. 189, 7507-7510 (2007).
- Ferandez, A., Hawkins, A. C., Summerfield, D. T., Harwood, C. S. Cluster II che genes from Pseudomonas aeruginosa are required for an optimal chemotactic response. J. Bacteriol. 184, 4374-4383 (2002).
- Cursino, L., et al. Identification of an Operon, Pil-Chp, That Controls Twitching Motility and Virulence in Xylella fastidiosa. Mol. Plant Microbe Interact. 10, 1198-1206 (2011).
- Hazelbauer, G. L., Falke, J. J., Parkinson, J. S. Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem. Sci. 33, 9-19 (2008).
- Costa, H. S., et al. Plant hosts of Xylella fastidiosa in and near southern California vineyards. Plant Dis. 88, 1255-1261 (2004).
- Shi, X. Y., Dumenyo, C. K., Hernandez-Martinez, R., Azad, H., Cooksey, D. A. Characterization of regulatory pathways in Xylella fastidiosa: genes and phenotypes controlled by algU. Appl. Environ. Microbiol. 73, 6748-6756 (2007).
- Matsumoto, A., Young, G. M., Igo, M. M. Chromosome-Based Genetic Complementation System for Xylella fastidiosa. Appl. Environ. Microbiol. 75, 1679-1687 (2009).
- Davis, M. J., Purcell, A. H., Thomson, S. V. Isolation Media for the Pierce’s Disease Bacterium. Phytopathology. 70, 425-429 (1980).
- Xia, Y. N., Whitesides, G. M. Soft lithography. Annu. Rev. Mater. Sci. 28, 153-184 (1998).
- Chaudhury, M. K., Whitesides, G. M. Direct measurement of interfacial interactions between semispherical lenses and flat sheets of poly-(dimethylsiloxane) and their chemical derivatives. Langmuir. 7, 1013-1025 (1991).
- Cruz, L. F., Parker, J. K., Cobine, P. A., De La Fuente, L. Calcium-enhanced twitching motility in Xylella fastidiosa is linked to a single PilY1 homolog. Appl. Environ. Microbiol. 80, 7176-7196 (2014).

