Time-lapse Confocal Imaging of Migrating Neurons in Organotypic Slice Culture of Embryonic Mouse Brain Using In Utero Electroporation
Summary
This protocol provides instructions for direct observation of radially migrating cortical neurons. In utero electroporation, organotypic slice culture, and time-lapse confocal imaging are combined to directly and dynamically study the effects of overexpression or downregulation of genes of interest in migrating neurons and to analyze their differentiation during development.
Abstract
In utero electroporation is a rapid and powerful approach to study the process of radial migration in the cerebral cortex of developing mouse embryos. It has helped to describe the different steps of radial migration and characterize the molecular mechanisms controlling this process. To directly and dynamically analyze migrating neurons they have to be traced over time. This protocol describes a workflow that combines in utero electroporation with organotypic slice culture and time-lapse confocal imaging, which allows for a direct examination and dynamic analysis of radially migrating cortical neurons. Furthermore, detailed characterization of migrating neurons, such as migration speed, speed profiles, as well as radial orientation changes, is possible. The method can easily be adapted to perform functional analyses of genes of interest in radially migrating cortical neurons by loss and gain of function as well as rescue experiments. Time-lapse imaging of migrating neurons is a state-of-the-art technique that once established is a potent tool to study the development of the cerebral cortex in mouse models of neuronal migration disorders.
Introduction
The neocortex is the major site of cognitive, emotional, and sensorimotor functions. It is composed of six horizontal layers oriented in parallel to the surface of the brain. During development progenitor cells in the lateral wall of the dorsal telencephalon give rise to projection neurons that migrate radially towards the pial surface and acquire a layer type-specific neuronal identity. After being generated in the ventricular/subventricular zones (VZ/SVZ) these neurons become transiently multipolar and slow their migration. After a short stay in the intermediate zone (IZ) they switch to a bipolar morphology, attach to the radial glial scaffold, and continue radially oriented migration into the cortical plate (CP). Upon reaching their final target projection neurons detach from the radial glial processes and acquire layer-specific identity. Mutations in genes affecting different steps of neuronal migration can cause severe cortical malformation, such as lissencephaly or white matter heterotopia1,2.
In utero electroporation is a rapid and powerful technique to transfect neural progenitor cells in the developing brain of rodent embryos3,4. With this technique it is possible to knockdown and/or overexpress genes of interest in order to study their functions in developing neurons. This method has specifically helped to describe the morphological details and characterize the molecular mechanisms of the process of radial migration5,6,7,8,9. Radially migrating neurons undergo dynamic changes in cell shape, migration speed, as well as migratory direction, which require direct and continuous observation over time. Organotypic slice culture and time-lapse confocal imaging of electroporated brains allow to directly observe migrating neurons over time. Using this combined approach, it is possible to analyze distinct features of migrating neurons that cannot be investigated in fixed tissue sections of electroporated brains.
We recently applied time-lapse confocal imaging of migrating neurons in slice cultures of electroporated brains to study the role of the transcription factor B cell CLL/lymphoma 11a (Bcl11a) during cortical development10. Bcl11a is expressed in young migrating cortical neurons and we used a conditional mutant Bcl11a allele (Bcl11aflox)11 to study its functions. Electroporation of Cre recombinase together with green fluorescent protein (GFP) into cortical progenitors of Bcl11aflox/flox brains allowed us to create a mosaic mutant situation, in which only few cells are mutated in an otherwise wild-type background. In this way, it was possible to study cell-autonomous functions of Bcl11a at the single cell level. We found that Bcl11a mutant neurons display reduced speed, shifts in their speed profiles, as well as random-like orientation changes during their migration10. In the outlined protocol we describe a workflow for successful electroporation and slice culture preparation12 of mouse brains, as well as time-lapse confocal imaging of cortical slice cultures.
Protocol
All experimental procedures were approved by the Animal Welfare Committee (Regierungspräsidium Tübingen) and carried out in accordance by the German Animal Welfare Act and the EU Directive 2010/63/EU.
1. In Utero Electroporation
- Microinjection needles
- Pull borosilicate glass capillaries (outer diameter: 1.0 mm, inner diameter: 0.58 mm, length: 100 mm) into microinjection needles using a micropipette puller with a box filament (2.5 mm x 2.5 mm) and the following program: HEAT: 540, PULL: 125, VELOCITY: 20, and DELAY: 140. Determine the HEAT value for every individual filament by performing a RAMP test (HEAT value: RAMP+25).
- Bevel needles at an angle of 38° and intermediate to high revolution speed with a microgrinder to obtain a tip size of 20 to 30 µm for injecting at embryonic day (E) 13.5 or younger and 30 to 40 µm for injecting at older embryonic stages. For storage, fix needles in a box or petri dish with modeling clay to prevent damage to the tips.
- Plasmid DNA Solution
- Prepare plasmid DNA construct containing fluorescent reporter protein (e.g. GFP) using an endotoxin-free maxi preparation kit according to the manufacturer's instructions.
- Dilute plasmid DNA solution to a final concentration of 1 – 2 µg/µL in endotoxin-free Tris-EDTA buffer containing Fast Green at a final concentration of 0.01%. Aliquot solution and store at -20 °C.
- Bacteriostatic Sodium Chloride Solution
- Prepare bacteriostatic sodium chloride solution by adding benzyl alcohol to a final concentration of 0.9% to isotonic 0.9% sodium chloride solution. Sterile filter solution immediately before use.
- Carprofen Injection Solution
- Prepare carprofen injection solution by adding carprofen to a final concentration of 0.5 mg/mL to sterile isotonic 0.9% sodium chloride solution. Store at 4 °C for up to 28 days.
- Anesthesia, DNA Injection, and Electroporation
- Weigh an E14.5 timed-pregnant mouse and place it into a transparent anesthetizing chamber connected to a vaporizer and saturated with 5% isoflurane. When the animal is unconscious, transfer it to an anesthetizing mask attached to a 37 °C warming plate and keep it anesthetized with 1.8 – 2.2% isoflurane.
Note: Assess the development of surgical anesthesia by loss of tail pinch and pedal reflexes (toe pinch). - For analgesia, inject 100 µL of carprofen injection solution per 10 mg of mouse bodyweight (5 mg/kg bodyweight) subcutaneously. Turn the mouse with its back down and carefully cover the eyes with petroleum jelly to prevent them from drying.
- Gently spread out the limbs and fix them to the warming plate with surgical tape. Sterilize the abdomen with 70% ethanol followed by iodine solution (7.5 mg/mL) using cellulose swabs. Repeat this procedure three times to ensure for proper disinfection.
- Cover the abdomen with sterile gauze in which an incision has been cut to spare out a field (diameter: 2 – 2.5 cm) for the abdominal incision. Moisten the gauze with bacteriostatic sodium chloride solution.
Caution: Reassess the development of surgical anesthesia by loss of the pedal reflex. - Using Micro Adson Forceps (serrated, length: 12 cm) and fine scissors (angled to side, length: 9 cm) cut the skin approximately 1.5 cm along the midline of the abdomen. Cut the underlying muscle along the linea alba.
- Remove one uterine horn with ring forceps (length: 9 cm, outer diameter: 3 mm, inner diameter: 2.2 mm) and gently place it onto the moistened gauze.
Caution: Hold the uterine horn with ring forceps only in between embryos and do not perturb the placentas or any supply vessels. Keep the uterus moist at all times with bacteriostatic sodium chloride solution. - Carefully position an embryo with ring forceps and locate the lateral ventricle, which presents as a crescent shaped shadow parallel to the sagittal sinus. The injection site lies approximately in the middle of a line between the pigmented eye and the confluence of sinuses, where the sagittal sinus branches to two transverse sinuses. Push the microinjection needle through the uterine wall and into the lateral ventricle.
Note: If necessary, the depth of the injection can be corrected by retraction of the needle. - Inject 1 to 2 µL of DNA solution into the lateral ventricle with a microinjector, which is operated with a foot pedal, using the following settings: 25 psi, 10 to 20 ms per pulse, and 5 to 10 pulses. Successful injection can be monitored by the colored DNA solution, which should fill the ventricular system.
- Place tweezers-type electrodes (3 mm diameter for E13.5 or younger and 5 mm diameter for E14.5 or older) in such a way that the 'positive' terminal is on the same side as the injected ventricle and the 'negative' terminal is on the opposite side of the injected ventricle below the ear of the embryo's head.
- Moisten the electroporation site with a few drops of bacteriostatic sodium chloride solution and apply 5 electrical current pulses (35 V for E13.5 or younger and 40 V for E14.5 or older) of 50 ms duration with 950 ms intervals.
Note: Currents of 60 to 90 mA are usually sufficient for successful electroporation. Lower currents will not efficiently transfect neurons, whereas higher currents may lead to embryonic death. - Repeat the procedure (c.f. Step 1.5.7. to 1.5.10.) for every embryo of the uterine horn and gently place it back into the abdominal cavity.
- Remove the uterine horn from the other side and repeat the procedure described in Step 1.5.7. to 1.5.11.
- Close the abdominal cavity by suturing the muscle layer before suturing the skin using Micro Adson Forceps, Mathieu Needle Holder (tungsten carbide, length: 14 cm), and non-absorbable surgical suture (3/8 circle, 13 mm, taper point).
Caution: Be careful not to capture the uterus or any other organs during suture. - Carefully disinfect the skin suture with iodine solution (7.5 mg/mL) and gently remove periocular excess of petroleum jelly using cellulose swabs. Place the mouse under an infrared lamp until it wakes up. Closely monitor the mouse during the next two days. Repeat the analgesia treatment as described in Step 1.5.2 after 12 to 24 h.
- Weigh an E14.5 timed-pregnant mouse and place it into a transparent anesthetizing chamber connected to a vaporizer and saturated with 5% isoflurane. When the animal is unconscious, transfer it to an anesthetizing mask attached to a 37 °C warming plate and keep it anesthetized with 1.8 – 2.2% isoflurane.
2. Organotypic Slice Culture
- Laminin Stock Solution
- Dissolve 1 mg of lyophilized laminin in sterile water to obtain a 1 mg/mL laminin stock solution. Prepare 50 µL aliquots and store at -80 °C.
- Poly-L-Lysine Stock Solution
- Dissolve 50 mg of poly-L-lysine in sterile water to obtain a 1 mg/mL poly-L-lysine stock solution. Prepare 0.5 mL aliquots and store at -20 °C.
- Complete Hank's Balanced Salt Solution (Complete HBSS)
- Prepare complete HBSS by combining 50 mL of 10x HBSS solution, 1.25 mL of 1 M HEPES buffer (pH 7.4), 15 mL of 1 M D-glucose solution, 5 mL of 100 mM calcium chloride solution, 5 mL of 100 mM magnesium sulfate solution, and 2 mL of 1 M sodium hydrogen carbonate solution. Add sterile water up to 0.5 L, sterile filter, and store at 4 °C.
- Slice Culture Medium
- Combine 35 mL of Basal Medium Eagle (BME), 12.9 mL complete HBSS (c.f. section 2.3.1.), 1.35 mL of 1 M D-glucose, 0.25 mL of 200 mM L-glutamine, and 0.5 mL of penicillin-streptomycin to obtain 50 mL of slice culture medium. Sterile filter and add horse serum to a final concentration of 5%. Store at 4 °C for up to 4 weeks.
- Low Melting Point (LMP) Agarose Solution
- Prepare 3% LMP agarose solution by dissolving 1.5 g of LMP agarose in 50 mL of complete HBSS by heating in a microwave oven for 1 to 2 min at intermediate power.
Note: Monitor heat carefully to prevent overflow during boiling. - Place solution in a water bath at 38 °C until ready for use. Store at 4 °C and reuse once.
- Prepare 3% LMP agarose solution by dissolving 1.5 g of LMP agarose in 50 mL of complete HBSS by heating in a microwave oven for 1 to 2 min at intermediate power.
- Coating of Membrane Inserts
- Add one aliquot of laminin stock solution (c.f. Step 2.1.1.) and one aliquot of poly-L-lysine stock solution (c.f. Step 2.2.1.) to 6 mL of sterile water to obtain coating solution (enough for 6 inserts).
- Place membrane inserts into a 6-well plate containing 2 mL of sterile water in the bottom of each well. Add 1 mL of coating solution (c.f. Step 2.6.1.) on top of each membrane and incubate overnight at 37 °C and 5% carbon dioxide atmosphere.
Note: Only use inserts with optically clear membranes, such as polytetrafluoroethylene (PTFE) membrane. - Wash the membranes three times with 1 mL of sterile water and dry. Use coated membranes on the same day or store in a clean 6-well plate at 4 °C for up to four weeks.
- Brain Dissection and Embedding
- Two days after the electroporation place the mouse into a transparent chamber connected to a vaporizer and saturated with 5% isoflurane. When the animal is unconscious, remove it from the chamber and sacrifice it by cervical dislocation.
- Remove the uterus containing the embryos and place it into a 10-cm Petri dish with ice-cold complete HBSS.
Note: From this point onward keep all solutions, embryos, and brains on ice. - Use a stereomicroscope, fine tipped forceps (straight, 11 cm) and a pair of Vannas Tübingen Spring Scissors (angled up, length: 9.5 cm) to separate each embryo from the uterus and transfer it to another petri dish containing ice-cold complete HBSS.
- Make an incision at the level of the brain stem and cut along the sagittal midline. Peel away the skin and cartilage covering the brain. Then, cut off the brain stem right behind the hemispheres and remove the brain from the skull. Transfer the brain with a micro-spoon spatula (185 mm x 5 mm) to a 12-well plate containing ice-cold complete HBSS. Collect all brains from the litter in the 12-well plate.
- Use a fluorescent stereomicroscope to screen the brains in the 12-well plate for fluorescence brightness as well as size of the electroporated region. Select 2 to 4 brains with bright fluorescent signals in the presumptive somatosensory cortex for further processing.
- Pour 3% LMP agarose solution kept at 38 °C into peel-away disposable embedding mold. Remove brain from the 12-well plate with a micro-spoon spatula and carefully drain excess complete HBSS around the brain using fine tissue paper.
- Place the brain gently into the agarose solution, push it to the bottom of the mold, and carefully adjust its position using a 20-gauge needle. For coronal sections, orient the brain with the olfactory bulbs pointing upwards. Keep the mold on ice until the agarose solution is solidified and continue with sectioning the brain.
- Vibratome Sectioning and Slice Culture
- Remove the agarose block from the mold and place it onto the lid of a sterile petri dish. Trim away excess agarose with a clean razor blade leaving approximately 2 to 3 mm below the olfactory bulbs and 1 mm of agarose on the other sides. Fix the trimmed agarose block with the olfactory bulbs pointing downwards to a specimen stage using cyanoacrylate glue.
Note: Sterilize instruments and equipment with 70% ethanol before sectioning. - Transfer the specimen stage to the slicing chamber of a vibrating blade microtome containing ice-cold complete HBSS and position the brain with its dorsal side towards the blade. Cut 250 µm thick brain slices containing the electroporated region with a low to moderate amplitude of 0.9 mm and a very slow sectioning speed of 0.09-0.12 mm/s. Usually 4 to 6 slices with bright fluorescent signal are obtained from each successfully electroporated brain.
- With a bent micro-spoon spatula and fine tipped forceps, carefully transfer the brain slices with the surrounding agarose to a 6-well plate containing ice-cold complete HBSS.
Caution: Be careful not to damage the brain tissue by only touching the agarose rim around the sections with the forceps during transfer. - Process all selected brains as described in Steps 2.8.1. to 2.8.3.
- Moisten the membrane inserts with 100 µL of complete HBSS before placing the brain slices onto the membranes to facilitate positioning of the slices. Up to 5 slices can be placed on 1 membrane insert.
- Carefully transfer the slices onto the membranes using a bent micro-spoon spatula and fine tipped forceps. Gently pull a slice onto the spatula by only touching the agarose rim with the forceps and gently push the slice from the spatula onto the membrane. Position the slice using forceps and continue to transfer the remaining slices. Remove excess complete HBSS with a pipette.
Note: Do not overlap the slices with one another. - With the help of forceps carefully place the membrane inserts with the slices into a 6-well plate containing 1.8 mL of slice culture medium (c.f. Step 2.4.1.) and incubate at 37 °C and 5% carbon dioxide atmosphere until time-lapse imaging.
- Remove the agarose block from the mold and place it onto the lid of a sterile petri dish. Trim away excess agarose with a clean razor blade leaving approximately 2 to 3 mm below the olfactory bulbs and 1 mm of agarose on the other sides. Fix the trimmed agarose block with the olfactory bulbs pointing downwards to a specimen stage using cyanoacrylate glue.
3. Time-Lapse Imaging
- Microscope Setup
- Set a climate chamber placed onto the stage of an inverted confocal microscope to 37 °C and 5% carbon dioxide atmosphere. Use a hybrid detector to reduce laser intensity and improve cell viability. For detailed analysis, use an extra-long working distance 40X dry objective with a numerical aperture of 0.6 or higher.
Note: All components of the microscope need to be pre-warmed to 37 °C for 2 – 3 h before imaging to provide a stable focus during the imaging process.
- Set a climate chamber placed onto the stage of an inverted confocal microscope to 37 °C and 5% carbon dioxide atmosphere. Use a hybrid detector to reduce laser intensity and improve cell viability. For detailed analysis, use an extra-long working distance 40X dry objective with a numerical aperture of 0.6 or higher.
- Slice Imaging
- Select a brain slice for imaging from the 6-well plate containing the membrane inserts using an inverted fluorescence microscope.
Caution: Avoid prolonged exposure time to ultraviolet light as this may cause cellular photodamage.
Note: Select a slice with bright single neurons in the upper SVZ migrating radially towards the pial surface of the slice. Fine fluorescent processes of radial glial cells that span the entire CP indicate an intact radial glial scaffold, which is used by radially migrating cortical neurons. - Transfer the membrane insert with the slice, which was selected for time-lapse imaging, into a 50 mm diameter glass bottom dish containing 2 ml of slice culture medium with the help of forceps. Place the dish into the climate chamber of the confocal microscope.
- Set the resolution to 512 by 512 pixels and increase the scan speed from 400 Hz to 700 Hz in order to increase the frame rate from about 1.4 to about 2.5 frames per second, respectively. Use no more than two times averaging. Define a z-stack through the electroporated region (usually 400-100 μm) with a step size of 1.5 μm. Collectively, these settings allow for a sufficient resolution and brightness of the image while keeping photodamage low during acquisition. Start the time-lapse series by taking a z-stack every 30 min.
Note: Prolonged exposure of the slice to laser light will induce photodamage reducing the viability of the cells. Adjust the exposure time to laser light accordingly. - Analyze time-lapse series using ImageJ software (https://imagej.nih.gov/iij/) or equivalent.
- Select a brain slice for imaging from the 6-well plate containing the membrane inserts using an inverted fluorescence microscope.
Representative Results
Previously, we have shown that genetic deletion of Bcl11a by in utero electroporation impairs radial migration of late-born upper-layer projection neurons10. Electroporation of a DNA plasmid vector containing Cre-IRES-GFP efficiently deleted Bcl11a in conditional Bcl11aflox/flox brains11. When we analyzed E14.5 electroporated brains three days after the electroporation, most control neurons had migrated into the CP, whereas many Bcl11a mutant neurons were stalled along the migratory route in the IZ and VZ/SVZ. Moreover, we found an increase in the number of multipolar cells at the expense of bipolar cells specifically in the IZ upon Bcl11a deletion suggesting defects in polarization10.
To directly and dynamically analyze migratory behavior of Bcl11a mutant neurons we used time-lapse confocal imaging of migrating neurons in organotypic slice culture prepared from electroporated brains. Overview time-lapse series using a 10X objective with a numerical aperture of 0.4 confirmed our results, that Bcl11a mutant projection neurons have defects in radial migration. Within 36 h many control neurons had migrated into the CP, whereas only few Bcl11a mutant neurons had left the IZ and migrated into the CP. Interestingly, it seemed that many Bcl11a mutant neurons did not migrate directly towards the pial surface of the brain, but slowed down migration and turned tangentially or even back towards the ventricle (Movie 1). To further analyze these findings we generated time-lapse series at a higher magnification using a 40x objective with a numerical aperture of 0.6. Our data show that Bcl11a mutant neurons failed to efficiently polarize and to continue radially oriented migration into the CP. Within 12 h, many Bcl11a mutant neurons failed to switch from a multipolar to a bipolar morphology and instead projected and retracted many processes into the surrounding environment (Movie 2, Figure 1A).
Furthermore, we traced the migration paths of individual neurons in different time-lapse series and used these to calculate specific parameters of migrating neurons. We found that Bcl11a mutant neurons repetitively undergo phases of several hour duration with reduced migration speed and random-like orientation changes (Movie 3; Figure 1B, red arrowheads). In particular, the speed profile of Bcl11a mutant neurons was shifted from higher speed (25 µm/h) towards lower speed (5 µm/h) in comparison to control neurons (Figure 1C). In line with this, the overall migration speed of Bcl11a mutant neurons was significantly reduced from 10.34 ± 0.34 µm/h in controls to 6 ± 0.54 µm/h (p = 2.4889E-05) (Figure 1D). Finally, the deviation from directed migration towards the pial surface of the brain was significantly increased from 17.15 ± 2.13° in control to 40.16 ± 4.42° in Bcl11a mutant neurons (p = 0.0002) (Figure 1E). Together these results demonstrate that time-lapse confocal imaging of GFP electroporated neurons in organotypic slice culture is a valuable method to study molecular mechanisms controlling the process of neuronal migration.
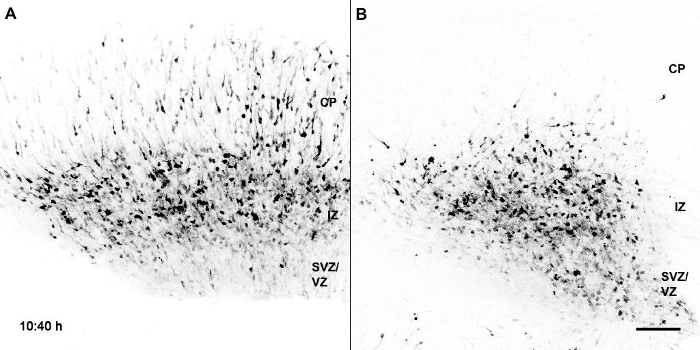
Movie 1: Migration of GFP Labeled Control in Comparison to Bcl11a Mutant Neurons in Cortical Slices. Bcl11aflox/flox brains were electroporated at E14.5 with a plasmid vector containing IRES-GFP (A) or Cre-IRES-GFP (B) and slice cultures were prepared at E16.5. VZ/SVZ; ventricular/subventricular zones; IZ, intermediate zone; CP, cortical plate. Scale bar = 100 µm. The movie is comprised of 108 frames at the rate of 5 frames/s. Reprinted from Wiegreffe et al.10 with permission from Elsevier. Please click here to view this video. (Right-click to download.)
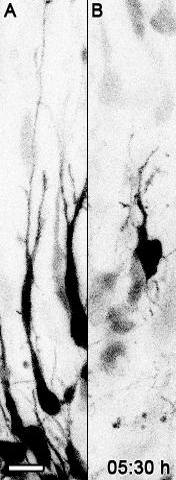
Movie 2: Polarization of a GFP Labeled Control in Comparison to Bcl11a Mutant Neurons in Cortical Slices. Bcl11aflox/flox brains were electroporated at E14.5 with a plasmid vector containing IRES-GFP (A) or Cre-IRES-GFP (B) and slice cultures were prepared at E16.5. Scale bar = 10 µm. The movie is comprised of 25 frames at the rate of 5 frames/s. Reprinted from Wiegreffe et al.10 with permission from Elsevier. Please click here to view this video. (Right-click to download.)
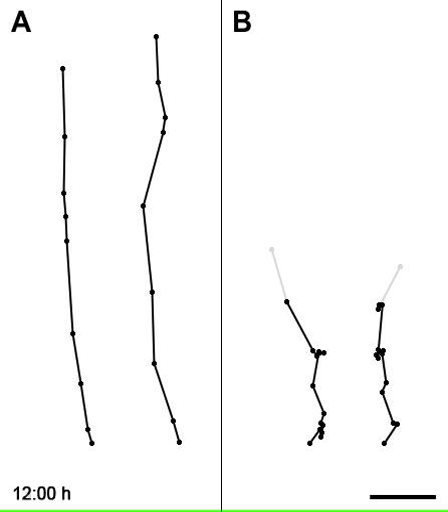
Movie 3: Animated Traces with 1 h Interval Resolution of Representative Control and Bcl11a Mutant Neurons. Traces of migrating neurons were obtained from time-lapse series of E16.5 slice cultures of Bcl11aflox/flox brains that were electroporated at E14.5 with a plasmid vector containing IRES-GFP (A) or Cre-IRES-GFP (B). Scale bar = 20 µm. Reprinted from Wiegreffe et al.10 with permission from Elsevier. Please click here to view this video. (Right-click to download.)
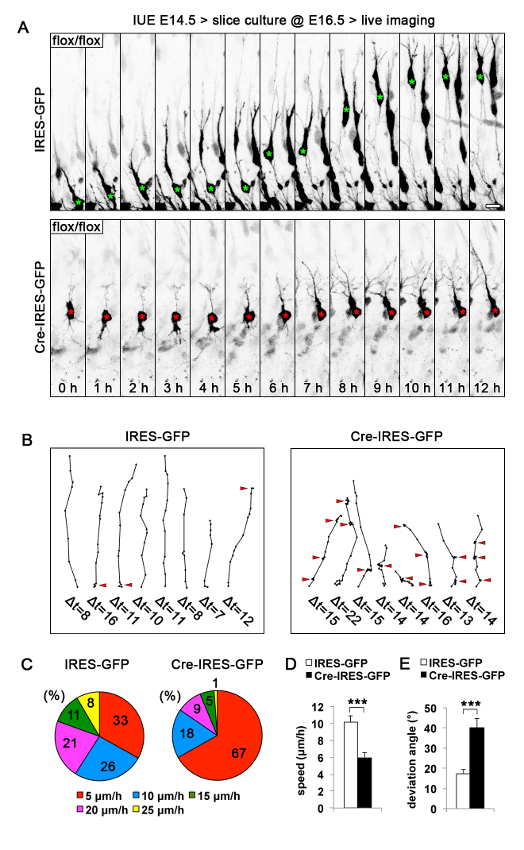
Figure 1: Migration Behavior of Bcl11a Mutant Upper-Layer Cortical Neurons.
(A) Representative images of migrating GFP positive neurons in E16.5 slice cultures from Bcl11aflox/flox brains electroporated at E14.5 with either Cre-IRES-GFP or a control vector over a total imaging period of 12 h. (B) Representative traces with 1 hour interval resolution of migrating GFP positive neurons in E16.5 slice cultures from Bcl11aflox/flox brains electroporated at E14.5 with Cre-IRES-GFP or a control vector over total imaging periods of up to 22 h. Bcl11a mutant neurons frequently undergo repetitive phases of reduced migration speed and randomly changed orientation (marked by red arrowheads). (C) Speed profiles calculated from traces of migrating neurons as shown in B. (D-E) Quantification of speed (D) and deviation angle from radial orientation (E) of migrating GFP positive neurons in E16.5 slice cultures from Bcl11aflox/flox brains electroporated at E14.5 with Cre-IRES-GFP or a control vector (n = 15). Mean ± s.e.m.; Student's t test; ***p <0.001. Scale bar = 10 μm. Reprinted from Wiegreffe et al.10 with permission from Elsevier. Please click here to view a larger version of this figure.
Discussion
Radial migration is a key process in neocortex development. Mutations in genes affecting different steps of this process can cause severe cortical malformations, including lissencephaly and white matter heterotopia1,2. We recently showed that Bcl11a, which is expressed in young migrating cortical projection neurons, plays a role in radial migration. We used time-lapse confocal imaging of migrating neurons in acute cortical slices of electroporated brains to directly demonstrate that genetic deletion of Bcl11a in migrating neurons causes polarization and migration defects. Time-lapse series were used to trace migration paths of individual neurons and calculate specific parameters of migrating neurons, including speed profiles, average migration speed, and migratory direction10.
This protocol describes an important approach when studying molecular mechanisms in radial migration. By using short hairpin RNA or DNA expression plasmids, almost any gene of interest can be knocked down or overexpressed, respectively. It is a powerful approach to combine electroporation with the Cre-LoxP system. Transfection of brains of conditional mutant ("floxed") mice with Cre recombinase generates a mosaic mutant situation and allows selective analysis of single mutant neurons that are surrounded by wild-type cells. In this way, cell autonomous molecular mechanisms in migrating neurons can be investigated. Also, functional rescue experiments are easily performed. Preparing time-lapse series of electroporated brain slices allows dynamically analyzing migratory behavior of single migrating neurons, which provides much more information than can be obtained from fixed brain sections. In utero electroporation3,4 requires initial training but – once mastered – is a rapid and straightforward technique to reproducibly transfect neurons of the cerebral cortex in vivo. In the protocol described here we used in utero electroporation at E14.5, when late-born upper-layer neocortical projection neurons are born. For the study of early-born deep-layer projection neurons in utero electroporation has to be performed on earlier developmental stage (i.e. E12.5)13. In principle, the protocol may also be used to study migration of other neuronal subpopulations in the brain that can be transfected with electroporation, including migration of interneurons14 and cerebellar granule cells15.
The slice culture protocol that we describe was adapted from Polleux and Gosh12 and previously, we have used it for ex utero electroporation16,17 to study hippocampal development. Most importantly, the brain slices have to be handled with care throughout the procedure to maintain their viability. We use an inverted microscope and optical quality glass bottom dishes to image the slice culture resting on a cell culture insert. This configuration is very easy to set up and allows sterile conditions, since the objective never gets in touch with the slice culture or medium. However, long working distance objectives (>2 mm) with a sufficiently high numerical aperture (0.6 or higher) are required. Other studies have put the brain slice directly onto the glass bottom and used a collagen matrix18,19 to fix the slice in place, which makes the use of objectives with a shorter free working distance possible. However, using cell culture inserts provides easy handling, reproducible results, and allows the researcher to image the slices even 1 to 2 days after their preparation without compromising viability of the brain slice (data not shown). Another critical issue concerns damaging the slices by laser light. We have used a highly sensitive hybrid detector, which allowed us to reduce laser power to a minimum while still obtaining sufficient fluorescent signals for time-lapse imaging. Using multiphoton imaging would further reduce photodamage and would allow for a deeper tissue penetration as well as more efficient light detection as compared to conventional confocal microscopy.
Despite its usefulness to study neuronal migration, the protocol has limitations and cannot entirely preserve the situation of migrating neurons in the intact brain. Extracellular matrix, the adhesive intercellular contacts, and the vasculature within the migratory zone can dynamically influence radially migrating neurons20,21. Specifically, upper layer neurons depend on elongated radial glial processes for their migration22. For successful imaging, it is therefore important to select brain slices with GFP electroporated radial glial cells that extend their processes through the entire cortical width. 'Oblique slices', in which the radial glial scaffold has been cut off before reaching the pial surface of the brain, should be discarded from further analysis. Sometimes, cell death may occur at the air-surface of the brain slice, which restricts image acquisition to deeper region within the sample. Electroporated neurons in cultured brain slices may migrate less efficiently compared to electroporated neurons in vivo, which can be caused by insufficient recovery from preparation. This requires analyses of multiple slice cultures for both experimental and control situations, and critical interpretation of representative data sets. In some cases the insults associated with in utero electroporation may cause embryonic death or structural alterations of the brain, including enlarged ventricles, asymmetrically thinned cortex, or a glial scar formation resulting from DNA injection. In those cases, the sample should be discarded from further analysis.
While brain slices from embryonic donors survive well on membrane inserts23, analyses are restricted to early developmental events, including neuronal migration, and we have not used it for studying later events, such as dendrite development or synaptogenesis. In summary, we provide a detailed protocol to directly and dynamically study neuronal migration of electroporated neurons in slice cultures, which can be readily adapted to analyze functions of genes involved in neurodevelopmental disorders.
Declarações
The authors have nothing to disclose.
Acknowledgements
We thank Jacqueline Andratschke, Elena Werle, Sachi Takenaka, and Matthias Toberer for excellent technical assistance, as well as Victor Tarabykin for helpful discussions. This work was supported by a grant of the Deutsche Forschungsgemeinschaft to S.B. (BR-2215).
Materials
| isoflurane | Abbott Laboratories | 506949 | Forene |
| 6-well plate | Corning | 351146 | |
| 12-well plate | Corning | 351143 | |
| non-absorbable surgical suture | Ethicon | K890H | 3/8 circle, 13 mm, taper point |
| Micro Adson Forceps | Fine Science Tools | 11018-12 | serrated, length: 12 cm |
| fine scissors | Fine Science Tools | 14063-09 | angled to side, length: 9 cm |
| Mathieu Needle Holder | Fine Science Tools | 12510-14 | tungsten carbide, length: 14 cm |
| fine tipped forceps | Fine Science Tools | 11370-40 | straight, 11 cm |
| Vannas Tübingen Spring Scissors | Fine Science Tools | 15005-08 | angled up, 9.5 cm |
| ring forceps | Fine Science Tools | 11103-09 | OD: 3mm, ID, 2.2 mm, length: 9 cm |
| HBSS (10X) | Gibco | 14180046 | |
| L-Glutamine | Gibco | 25030081 | |
| Penicillin/Streptomycin | Gibco | 15140122 | |
| horse serum | Gibco | 26050088 | |
| BME | Gibco | 41010026 | |
| borosilicate glass capillaries | Harvard Apparatus | 30-0016 | 1.0 OD x 0.58 ID x 100 L mm |
| anesthsesia system | Harvard Apparaus | 72-6471 | |
| anesthetizing chamber | Harvard Apparaus | 34-0460 | |
| fluosorber filter canister | Harvard Apparaus | 34-0415 | |
| low melting point agarose | Invitrogen | 16520100 | |
| vibrating blade microtome | Leica | VT1200 S | |
| fluorescence stereo microscope | Leica | M205 FA | |
| stereo microscope | Leica | M125 | |
| inverted fluorescence tissue culture microscope | Leica | DM IL LED | |
| confocal laser scanning microscope | Leica | TCS SP5II | |
| hybrid detector | Leica | HyD | |
| objective, 40x/0.60 NA | Leica | 11506201 | |
| microscope temperature control system | Life Imaging Services | Cube, Brick & Box | |
| cell culture insert | Millipore | PICM0RG50 | |
| microgrinder | Narishige | EG-45 | use 38° angle for beveling |
| microinjector | Parker Hannifin | 052-0500-900 | Picospritzer III |
| carprofen | Pfizer Animal Health | NDC 61106-8507 | Rimadyl |
| emdedding mold | Polysciences | 18986-1 | |
| endotoxin-free plasmid maxi kit | Qiagen | 12362 | |
| fast green | Sigma | F7252 | |
| laminin | Sigma | L2020 | |
| poly-L-lysine | Sigma | P5899 | |
| HEPES | Sigma | H4034 | |
| D-glucose | Sigma | G6152 | |
| calcium chloride | Sigma | C7902 | |
| magensium sulfate | Sigma | M2643 | |
| sodium bicarbonate | Sigma | S6297 | |
| square wave electroporator | Sonidel | CUY21EDIT | |
| tweezers with 5 mm platinum disk electrodes | Sonidel | CUY650P5 | |
| micropipette puller | Sutter Instrument | P-97 | |
| box filament | Sutter Instrument | FB255B | 2.5 mm x 2.5 mm |
| micro-spoon spatula | VWR | 231-0191 | 185 mm x 5 mm |
| glass bottom dish, 50 mm | World Precision Instruments | FD5040-100 |
Referências
- Evsyukova, I., Plestant, C., Anton, E. S. Integrative mechanisms of oriented neuronal migration in the developing brain. Annu Rev Cell Dev Biol. 29, 299-353 (2013).
- Kwan, K. Y., Sestan, N., Anton, E. S. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development. 139 (9), 1535-1546 (2012).
- Saito, T., Nakatsuji, N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 240 (1), 237-246 (2001).
- Tabata, H., Nakajima, K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neurociência. 103 (4), 865-872 (2001).
- LoTurco, J. J., Bai, J. The multipolar stage and disruptions in neuronal migration. Trends Neurosci. 29 (7), 407-413 (2006).
- Noctor, S. C., Martinez-Cerdeno, V., Ivic, L., Kriegstein, A. R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 7 (2), 136-144 (2004).
- Tabata, H., Nakajima, K. Multipolar migration: the third mode of radial neuronal migration in the developing cerebral cortex. J Neurosci. 23 (31), 9996-10001 (2003).
- Pacary, E., et al. Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signaling. Neuron. 69 (6), 1069-1084 (2011).
- Tabata, H., Nagata, K. Decoding the molecular mechanisms of neuronal migration using in utero electroporation. Medical Molecular Morphology. 49 (2), 63-75 (2016).
- Wiegreffe, C., et al. Bcl11a (Ctip1) Controls Migration of Cortical Projection Neurons through Regulation of Sema3c. Neuron. 87 (2), 311-325 (2015).
- John, A., et al. Bcl11a is required for neuronal morphogenesis and sensory circuit formation in dorsal spinal cord development. Development. 139 (10), 1831-1841 (2012).
- Polleux, F., Ghosh, A. The slice overlay assay: a versatile tool to study the influence of extracellular signals on neuronal development. Sci STKE. (136), pl9 (2002).
- Greig, L. C., Woodworth, M. B., Galazo, M. J., Padmanabhan, H., Macklis, J. D. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 14 (11), 755-769 (2013).
- De Marco Garcia, N. V., Fishell, G. Subtype-selective electroporation of cortical interneurons. J Vis Exp. (90), e51518 (2014).
- Holubowska, A., Mukherjee, C., Vadhvani, M., Stegmuller, J. Genetic manipulation of cerebellar granule neurons in vitro and in vivo to study neuronal morphology and migration. J Vis Exp. (85), (2014).
- Venkataramanappa, S., Simon, R., Britsch, S. Ex utero electroporation and organotypic slice culture of mouse hippocampal tissue. J Vis Exp. (97), (2015).
- Simon, R., et al. A dual function of Bcl11b/Ctip2 in hippocampal neurogenesis. EMBO J. 31 (13), 2922-2936 (2012).
- Youn, Y. H., Pramparo, T., Hirotsune, S., Wynshaw-Boris, A. Distinct dose-dependent cortical neuronal migration and neurite extension defects in Lis1 and Ndel1 mutant mice. J Neurosci. 29 (49), 15520-15530 (2009).
- Nadarajah, B., Brunstrom, J. E., Grutzendler, J., Wong, R. O., Pearlman, A. L. Two modes of radial migration in early development of the cerebral cortex. Nat Neurosci. 4 (2), 143-150 (2001).
- Higginbotham, H., Yokota, Y., Anton, E. S. Strategies for analyzing neuronal progenitor development and neuronal migration in the developing cerebral cortex. Cereb Cortex. 21 (7), 1465-1474 (2011).
- Stubbs, D., et al. Neurovascular congruence during cerebral cortical development. Cereb Cortex. 19, i32-i41 (2009).
- Ayala, R., Shu, T., Tsai, L. H. Trekking across the brain: the journey of neuronal migration. Cell. 128 (1), 29-43 (2007).
- Humpel, C. Organotypic brain slice cultures: A review. Neurociência. 305, 86-98 (2015).

