In Vitro Differentiation of Mouse Granulocyte-macrophage-colony-stimulating Factor (GM-CSF)-producing T Helper (THGM) Cells
Summary
Here, we present a protocol to differentiate murine granulocyte-macrophage-colony-stimulating-factor-producing T helper (THGM) cells from naive CD4+ T cells, including isolation of naive CD4+ T cells, differentiation of THGM, and analysis of differentiated THGM cells. This method can be applied to studies of the regulation and function of THGM cells.
Abstract
The granulocyte-macrophage-colony-stimulating factor (GM-CSF)-producing T helper (THGM) cell is a newly identified T helper cell subset that predominantly secretes GM-CSF without producing interferon (IFN)γ or interleukin (IL)-17 and is found to play an essential role in the autoimmune neuroinflammation. A method of isolation of naive CD4+ T cells from a single-cell suspension of splenocytes and THGM cell generation from naive CD4+ T cells would be a useful technique in the study of T cell-mediated immunity and autoimmune diseases. Here we describe a method that differentiates mouse naive CD4+ T cells into THGM cells promoted by IL-7. The outcome of the differentiation was assessed by the analysis of the cytokines expression using different techniques, including intracellular cytokine staining combined with flow cytometry, a quantitative real-time polymerase chain reaction (PCR), and enzyme-linked immunosorbent assays (ELISA). Using the THGM differentiation protocol as described here, about 55% of the cells expressed GM-CSF with a minimal expression of IFNα or IL-17. The predominant expression of GM-CSF by THGM cells was further confirmed by the analysis of the expression of GM-CSF, IFNα, and IL-17 at both mRNA and protein levels. Thus, this method can be used to differentiate naive CD4+ T cells to THGM cells in vitro, which will be useful in the study of THGM cell biology.
Introduction
CD4+ T helper (TH) cells are essential components of the immune system, having crucial roles in the host defense against microbial pathogens, in cancer surveillance, and in autoimmunity1,2,3. Upon T cell receptor (TCR) activation, naive CD4+ T cells can be differentiated into TH1, TH2, TH 17, or regulatory T (Treg) cells under the influence of different cytokine milieus2,4,5. Recently, a new subset of TH cells, which predominantly produces GM-CSF, was identified and named THGM6. The differentiation of THGM cells is driven by IL-7 through the activation of a signal transducer and an activator of transcription 5 (STAT5). These cells express a large amount of GM-CSF while having a low expression of other TH-cell signature cytokines such as IFNγ and IL-176. GM-CSF was found to play a critical role in the development of CD4+ T cell-mediated neuroinflammation7,8. Compared to IFNγ- or IL-17-expressing autoreactive T cells, GM-CSF-expressing T cells transferred to wild-type (WT) mice caused an earlier disease onset and higher disease severity. In addition, Csf2-/- T cells failed to induce experimental autoimmune encephalomyelitis (EAE) after being adoptively transferred to WT recipients, whereas T cells lacking IFNγ or IL-17A retained the ability to mediate EAE7. Moreover, a blockade of GM-CSF using neutralizing antibodies ameliorated EAE disease severity8. Furthermore, a deficiency of STAT5 in T cells in mice resulted in a diminished THGM generation and, hence, in a resistance of the mice to EAE development6. These findings underscore the importance of GM-CSF-expressing TH cells in autoimmune neuroinflammatory disease. Thus, establishing a method to differentiate GM-CSF-expressing TH cells from naive CD4+ T cells would be important in the study of the pathogenesis of autoimmune neuroinflammation and T cell-mediated immune responses. However, a protocol that efficiently generates THGM cells from murine naive CD4+ has not been established.
Here we present a method that differentiates murine THGM cells from naive CD4+ T cells. This protocol describes the whole procedure, including the extraction of the spleen from the mouse, the preparation of a single-cell suspension, the CD4 positive selection, the fluorescence-activated cell sorting (FACS), and the TH cell differentiation and analysis. The differentiated T helper cells are analyzed by intracellular cytokine staining combined with flow cytometry to determine the cytokine expression at the single-cell level, by a quantitative real-time PCR to determine the cytokine expression at mRNA level, and by ELISA to assess the cytokine expression at protein level. This method can be applied to studies of THGM cell biology under various conditions, such as EAE, where GM-CSF plays an important role in pathogenesis.
Protocol
All mice used in this protocol were on the C57BL/6 genetic background and housed under specific pathogen-free conditions at the National University of Singapore. All experiments were performed using protocols approved by the Institutional Animal Care and Use Committee of the National University of Singapore.
1. Reagent and Material Preparation
- Prepare 500 mL of phosphate-buffered saline (PBS) containing 2% fetal bovine serum (FBS) and 1 mM ethylenediaminetetraacetic acid (EDTA).
NOTE: Keep the buffer on ice throughout the whole procedure for optimal results. - Prepare 50 mL of complete RPMI media containing RPMI 1640, 10% FBS, and 1% penicillin-streptomycin. Use complete RPMI media containing 50 µM β-mercaptoethanol for the THGM cell differentiation. Add the β-mercaptoethanol in a fume hood.
- Prepare 7.5 mL of an anti-CD3e antibody mixture by diluting anti-CD3e concentrated stock to 3 μg/mL in PBS. Coat 48-well plates by adding 150 μL of the antibody mixture to each well and incubate the plate at 37 °C for at least 1 h, or at 4 °C overnight.
- Sterilize a pair of scissors and forceps and keep them in 70% ethanol between dissections.
2. Preparation of Murine Splenocytes
- Euthanize the mouse (of 6–8 weeks old) using an institutionally-approved CO2 asphyxiation or cervical dislocation. Spray the mouse with 70% ethanol and mount it on a polystyrene block on its back. Transfer the mouse to a biological safety cabinet to proceed with the following steps.
- Hold the skin using a pair of forceps and cut the skin below the rib cage on the left side for about 4 cm, using a pair of scissors. Open the peritoneal sac to expose the spleen and use sterile forceps to extract the spleen.
- Place a 70 μm cell strainer on a 50 mL tube and pre-wet it with 2 mL of buffer. Place the spleens on the cell strainer (2–3 spleens for one cell strainer) and macerate them using the end of a 5 mL syringe plunger.
- Rinse the strainer and tube several times with 1–2 mL of cell isolation buffer until all cells are flushed into the tube. Discard the cell strainer and centrifuge the cells at 350 x g for 5 min at 4 °C. Discard the supernatant.
- Resuspend the cell pellet with 5 mL of cold (4 °C) ammonium-chloride-potassium (ACK) lysing buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM Na2EDTA; pH 7.2–7.4) and gently mix them for 2 min. Add 10 mL of complete RPMI media to neutralize the ACK buffer and immediately centrifuge the cells at 350 x g for 5 min at 4 °C. Discard the supernatant after the centrifugation.
3. Isolation of CD4+ T Cells Using Magnetic CD4 Microbeads and a Cell Separation Column
- Resuspend the cell pellet in 5 mL of cell isolation buffer. Filter the cell suspension using a pre-separation filter (30 μm) into a new 15-mL tube to remove any debris.
- Take 10 μL of the cell suspension and make a 10x dilution by adding 90 μL of buffer. Take 10 μL of the diluted cell suspension and mix it with 10 μL of trypan blue for cell counting. Count the cells in a hemocytometer to determine the yield of viable cells.
- Centrifuge the cells at 350 x g for 5 min at 4 °C and discard the supernatant.
- Resuspend the cells in 90 μL of buffer per 107 cells. Add 10 μL of magnetic CD4 microbeads per 107 cells. Incubate the cells for 15 min at 4 °C. For optimal results, mix the cell suspension gently every 5 min during incubation.
- While the cells are incubating, place a separation column on the magnetic stand. Pre-wet the column with 2 mL of buffer.
- At the end of the cell/bead incubation, wash the cells with 5 mL of buffer and centrifuge them at 350 x g for 5 min at 4 °C. Discard the supernatant.
- Resuspend the cells in 1 mL of buffer, load the sample into the cell separation column, and let it flow through. Use a 15-mL centrifuge tube to collect the CD4- cell fraction. Add 1 mL of buffer to wash the reservoir before it is dry. Repeat the washing step 2x.
- Remove the cell separation column from the magnetic stand and place it on a new 15 mL centrifuge tube. Add 2 mL of cell isolation buffer to the cell separation column and apply the plunger firmly to force the cells out of the column.
- Centrifuge the fraction at 350 x g for 5 min at 4 °C to obtain CD4+ cells. Discard the supernatant.
4. Purification of the Naive CD4+ T cells (CD4+CD25–CD44loCD62Lhi) by Fluorescence-activated Cell Sorting and THGM differentiation
- Resuspend the obtained CD4+ cell pellet in 500 µL of buffer. Mix the cells with a fluorescent-conjugated antibodies mixture containing CD4-PerCp (2.8 µg/mL), CD44-APC (2.4 µg/mL), CD25-PE (2 µg/mL), and CD62L-FITC (10 µg/mL) (Table 1). Incubate the cells on ice for 20–30 min, while protecting them from light.
NOTE: The concentrations of the antibodies were optimized previously in the lab. The number of antibodies listed is for cells from 1–2 mice. - After the incubation, wash the cells with 5 mL of buffer and centrifuge the cells at 350 x g for 5 min at 4 °C.
- Discard the supernatant and resuspend the cells in 500 µL of buffer. Filter the cells again using a nylon mesh.
- Transfer the cell suspension to a FACS tube for cell sorting. Precoat the collection FACS tubes with 500 µL of buffer.
- Obtain a CD4+CD25–CD44loCD62Lhi population (of naive CD4+ T cells) by FACS sorting. Gate on CD4+CD25– first, and then select CD44loCD62Lhi cells from this population.
- Collect the naive CD4+ T cell fractions into a new 15-mL centrifuge tube. Centrifuge the cells at 350 x g for 5 min at 4 °C and discard the supernatant.
- Resuspend the naive CD4+ T cells at a concentration of 106 cells/mL in complete RPMI media containing 50 µM β-mercaptoethanol.
- Aspirate the anti-CD3e antibodies used for precoating in the 48-well plate. Seed 0.25 million (250 µL) cells in each well with IL-7 (2 ng/mL), anti-CD28 (1 µg/mL), and anti-IFNγ (10 µg/mL) (Table 1).
NOTE: The concentrations of cytokine and antibodies were optimized previously in the lab for a THGM cell differentiation. - Incubate the cells at 37 °C with 5% CO2 for 3 days. Do not change the medium during the incubation.
5. Analysis of the Mouse THGM Cells Generated In Vitro
- Using a microscope, check the cell differentiation 3 d after the differentiation. Harvest the cells and centrifuge them at 350 x g for 5 min at 4 °C. Wash the cells 2x with complete RPMI media.
NOTE: Differentiated cells are larger in size than naive CD4+ T cells. - Resuspend the cells in 1 mL of complete RPMI media. Take 10 μL of cell suspension and mix it well with 10 μL of trypan blue to determine the cell number with a hemocytometer. Divide differentiated cells into three potions for a restimulation and analysis.
NOTE: Intracellular cytokine staining and ELISA assays require ≥0.5 million cells, and qPCR analysis requires ≥ 1 million cells. - For intracellular cytokine staining, restimulate the cells with phorbol 12-myristate 13-acetate (PMA) (100 ng/mL) and ionomycin (1 μg/mL) in the presence of a protein transport inhibitor (1 µL/mL) for 4–6 h.
- Harvest the cells and stain them with anti-CD4 antibody (PerCP-conjugated, 0.2 mg/mL, 1:100 dilution; or FITC-conjugated, 0.5 mg/mL, 1:100 dilution) for 30 min.
- Fix the cells with 200 μL of fixation buffer for 20–60 min. After the fixation, wash the cells 2x with 1 mL of permeabilization buffer.
- Perform intracellular staining with antibodies against GM-CSF (PE-conjugated, 0.2 mg/mL, 1:100 dilution), IL-17A (FITC-conjugated, 0.5 mg/mL, 1:100 dilution; APC-conjugated, 0.2 mg/mL, 1:100 dilution), IL-4 (APC-conjugated, 0.2 mg/mL, 1:100 dilution), and IFNγ (APC-conjugated, 0.2 mg/mL, 1:100 dilution; PE-conjugated, 0.2 mg/mL, 1:100 dilution) for 30 min in the dark.
- For a qPCR analysis of the cytokine gene expression by differentiated cells, activate the cells with plate-bound anti-CD3e (3 μg/mL) (precoating the plate as described in step 1.3). At 3 h after the stimulation, harvest the cells to isolate total RNA using a phenol RNA extraction reagent to prepare cDNA for the qPCR. The primers and program used for the qPCR are listed in Tables 2 and 3, respectively.
- For the analysis of the cytokine protein secretion by differentiated cells, restimulate the cells with plate-bound anti-CD3e (3 μg/mL) for 24 h (precoating the plate as described in step 1.3). Harvest the cell culture supernatant for IL-17 and GM-CSF ELISA to determine their concentrations.
Representative Results
Naive CD4+ T cells isolated from two 8-week-old male C57BL/6 mice were divided into three portions. One portion of the cells was differentiated into THGM cells following the protocol described. Another portion was cultured under a THGM condition in the presence of anti-IL-4 antibody (10 µg/mL) to test the influence of an IL-4 blockade in the differentiation of THGM. The last portion was cultured under a TH17 differentiation condition (3 µg/mL anti-CD3e, 1 µg/mL anti-CD28, 10 ng/mL TGFβ, 30 ng/mL IL-6, 10 µg/mL anti-IFNγ, and 10 µg/mL anti-IL-4). After 3 days of differentiation, the cells were harvested and restimulated to analyze the cytokine expression by intracellular cytokine staining, qPCR, and ELISA. Results from the intracellular cytokine staining and FACS analysis demonstrated that about 55% of the cells cultured under the THGM differentiation condition were GM-CSF-expressing cells (Figure 1A), whereas only about 2% of the cells differentiated under the TH17 condition expressed GM-CSF. In addition, compared to the TH17 differentiation condition which generated 8.17% IL-17-producing cells, the THGM differentiation condition only resulted in about 1% of IL-17-expressing cells. Under the two differentiation conditions tested, only a small fraction of the cells expressed IFNγ (<1%). Furthermore, few IL-4-expressing T cells (<1%) were seen in THGM or TH17 culture (Figure 1B). These results showed that using the described THGM cell differentiation condition, we have successfully generated T helper cells that predominantly express GM-CSF.
The successful differentiation of THGM cells was further confirmed by results from qPCRs and ELISA assays to examine the expression of GM-CSF and IL-17 at both RNA and protein levels in the differentiated cells. The generated THGM cells had a significantly higher expression of Csf2 but a much lower expression of Il17 compared to the TH17 cells (Figure 2A). As shown in Figure 2B, GM-CSF protein was detected in culture supernatants from both THGM and TH17 cells. Nevertheless, the GM-CSF concentration in the THGM culture supernatant was about threefold of that in TH17 cells. In addition, proteins of IL-17 (Figure 2B), IFNγ, and IL-4 were undetectable in the THGM cell culture supernatant. Since RORγt has been identified as the master transcription factor of TH17 cells, while STAT3 and STAT5 have been shown to have a great importance in the development of TH17 and THGM cells, respectively, we examined their mRNA expression in the THGM and TH17 cells. It was found that the THGM cells had a significantly lower expression of Rorc and a higher Stat5 expression than the TH17 cells (Figure 3). Interestingly, the THGM cells had a slightly lower (but not significant) expression of the Stat3 gene compared to the TH17 cells. These results indicated that although the THGM and the TH17 differentiation are governed by different transcriptional factors, they may share some common features.
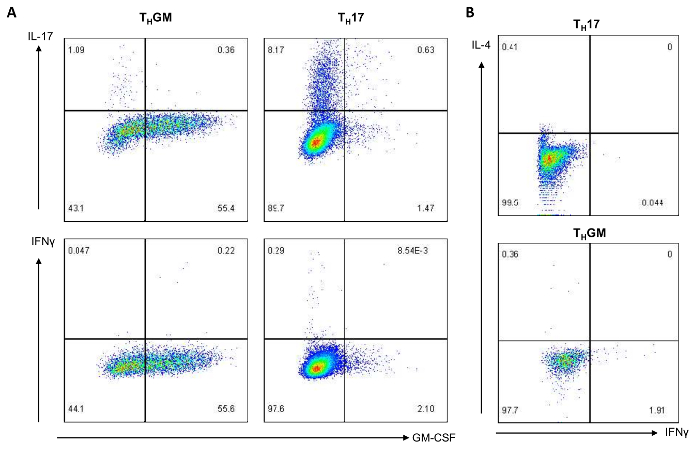
Figure 1: THGM cells predominantly express GM-CSF. THGM and TH17 cells were harvested on day 3 after the differentiation. (A) For 5 h, the cells were restimulated with PMA and ionomycin in the presence of a protein transport inhibitor. The expression of GM-CSF, IL-17, and IFNγ was analyzed by intracellular cytokine staining followed by flow cytometry. (B) The IL-4 and IFNγ expression by TH17 or THGM cells was analyzed. Please click here to view a larger version of this figure.
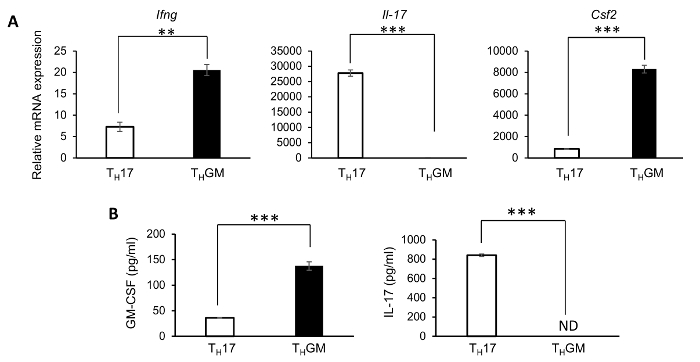
Figure 2: Expression of IL-17, IFNγ, and GM-CSF in TH17 and THGM cells. THGM and TH17 cells were harvested and restimulated with plate-bound anti-CD3e, 3 days after the differentiation. (A) Cells were harvested to isolate RNA for cDNA preparation, 3 h after the stimulation. The expressions of Ifng, Il-17, and Csf2 were determined by a quantitative real-time PCR (qPCR). (B) The culture supernatant was harvested at 24 h after the stimulation, to determine the concentration of GM-CSF in THGM cells, or IL-17 in TH17 cells, by ELISA. (** p <0.01, *** p <0.001.) Please click here to view a larger version of this figure.
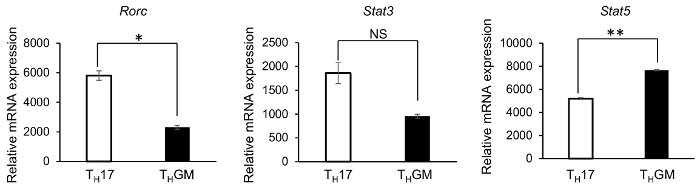
Figure 3: Rorc, Stat3, and Stat5 gene expression in TH17 and THGM cells. Differentiated THGM and TH17 cells were harvested and restimulated with plate-bound anti-CD3e for 3 h. Total RNA was isolated to prepare cDNA. The Rorc, Stat3, and Stat5 expressions were determined by qPCR. (* p <0.05, ** p <0.01; NS = not significant.) Please click here to view a larger version of this figure.
| Name | Stock concentration | working concentration | Dilution | volume in 500 μL staining buffer |
| CD4-PerCp | 0.2 mg/mL | 2.8 μg/mL | 1:71 | 7 μL |
| CD44-APC | 0.2 mg/mL | 2.4 μg/mL | 1:83 | 6 μL |
| CD25-PE | 0.2 mg/mL | 2 μg/mL | 1:100 | 5 μL |
| CD62L-FITC | 0.5 mg/mL | 10 μg/mL | 1:50 | 10 μL |
| GM-CSF-PE | 0.2 mg/mL | 2 μg/mL | 1:100 | |
| IL-17A-FITC | 0.5 mg/mL | 5 μg/mL | 1:100 | |
| IL-17A-APC | 0.2 mg/mL | 2 μg/mL | 1:100 | |
| IFNγ-APC | 0.2 mg/mL | 2 μg/mL | 1:100 | |
| IFNγ-PE | 0.2 mg/mL | 2 μg/mL | 1:100 | |
| IL-4-APC | 0.2 mg/mL | 2 μg/mL | 1:100 | |
| CD4-FITC | 0.5 mg/mL | 5 μg/mL | 1:100 | |
| IL-7 | 20 μg/mL | 2 ng/mL | 1:10000 | |
| Anti-CD28 | 0.5 mg/mL | 1 μg/mL | 1:500 | |
| Anti-IFNγ | 1 mg/mL | 10 μg/mL | 1:100 |
Table 1: Used cytokines and antibodies.
| Primer | Sequence | |
| Ifng Forward | 5’-TCAAGTGGCATAGATGTGGAAGAA-3’ | |
| Ifng Reverse | 5’-TGGCTCTGCAGGATTTTCATG-3’ | |
| Il-17 Forward | 5’-CTCCAGAAGGCCCTCAGACTAC-3’ | |
| Il-17 Reverse | 5’-AGCTTTCCCTCCGCATTGACACAG-3’ | |
| Csf2 Forward | 5’-TTTACTTTTCCTGGGCATTG-3’ | |
| Csf2 Reverse | 5’-TAGCTGGCTGTCATGTTCAA-3’ | |
| Rorc Forward | 5’-TTTGGAACTGGCTTTCCATC-3’ | |
| Rorc Reverse | 5’-AAGATCTGCAGCTTTTCCACA-3’ | |
| Stat3 Forward | 5’-TGGCCCTTTGGAATGAAGGGTACA-3’ | |
| Stat3 Reverse | 5’-CACTGATGTCCTTTTCCACCCAAGT-3’ | |
| Stat5 Forward | 5’-TGCCCGGCTGGAACTACACCTT-3’ | |
| Stat5 Reverse | 5’-ATGCCCCCGATTTCCGAGTCAC-3’ | |
Table 2: Primers for qPCR.
| step | Temp. (°C) | time | repeat | |
| 1 | 95 | 2 min | ||
| 2 | 95 | 3 s | ||
| 3 | 60 | 30 s | step 2 and 3, 39 times | read plate |
| 4 | 65–95 | 5 s | step 4, 30 times, +0.5°C each repeat | read plate |
Table 3: PCR program to assess gene expression.
Discussion
Here we described a protocol of an in vitro THGM differentiation from mouse naive CD4+ cells, followed by an analysis of the differentiated cells to validate the method. Of note, both spleen and lymph nodes can be used for naive CD4+ T cell purification and THGM differentiation. The cytokine expression determined by intracellular cytokine staining combined with flow cytometry showed that about 55% of the cells were induced to become GM-CSF-expressing cells under the THGM condition (Figure 1A). Compared to cells under the TH17 differentiation condition, the cells that differentiated under the THGM condition contained a background level of an IL-17-expressing population; however, the il17 gene expression is undetectable by qPCR (Figure 2A).
Interestingly, despite the addition of an IFNγ neutralizing antibody, we detected a low level of Ifng mRNA expression in both the TH17 and the THGM cells, and less than 1% of the THGM cells were IFNγ-expressing cells. (Figures 1 – 2). This could be due to the insufficient amount of IFNγ-neutralizing antibody used in the THGM condition, or to a possible contamination of innate immune cells (it is almost impossible to obtain 100% pure naive CD4+ T cells) that provided a trace amount of IL-12 for the possible TH1 cell generation. It is also possible that the THGM condition we used was not able to completely shut off the transcription of Ifng. We will address this issue in future. Nevertheless, IFNγ protein was not detected in the culture supernatants of THGM or TH17 by ELISA.
In the protocol presented here, we only used an IFNγ-blocking antibody together with IL-7 for the THGM differentiation. After 3 days of differentiation under this condition, more than 50% of the cells expressed GM-CSF, but not IL-17, IL-4, or IFNγ (Figure 1). In addition, the expression of Rorc, the gene that encodes RORγt, which is critical for the Th17 differentiation9, was significantly lower in the THGM cells compared to that in the TH17 cells (Figure 3). This protocol is, therefore, a more efficient method for the generation of GM-CSF-expressing T cells than another reported previously10. Our results also demonstrated that, in addition to the purity of naive T cells and IL-7 signaling, the prevention of the IFNγ-expressing T cell differentiation is a key for THGM generation.
The predominant expression of the GM-CSF cells that were generated using the described protocol demonstrated the successful differentiation of THGM. We would like to point out that the quality of cytokines and antibodies from different companies or even of different batches from the same company may not be the same. Therefore, we recommend that the number of cytokines and antibodies used in T helper cell differentiation should be tested in order to find out the optimal condition.
In summary, THGM cells that were generated using this protocol express a minimal level of IL-17, IL-4, or IFNγ. We are confident that this protocol works well in generating GM-CSF-expressing THGM cells in vitro. This method can be used to further study the biology of THGM cells in various conditions such as autoimmune neuroinflammation, including experimental autoimmune encephalomyelitis (EAE) in mice and human multiple sclerosis, where GM-CSF plays an important role in the pathogenesis11.
Declarações
The authors have nothing to disclose.
Acknowledgements
This study was supported by grants from the National University Health System of Singapore (T1-2014 Oct-12 and T1-2015 Sep-10).
Materials
| RPMI 1640 | Biowest | L0500-500 | |
| FBS Heat Inactivated | Capricorn | FBS-HI-12A | |
| Penicillin-Streptomycin | Gibco | 15140122 | |
| 10x PBS | 1st Base | BUF-2040-10 | Diluted to 1x PBS in sterile water |
| EDTA | 1st Base | BUF-1053-100ml-pH8.0 | |
| 10X ACK buffer | Biolegend | 420301 | diluted in sterile water |
| cell strainer | SPL Life Sciences | 93070 | |
| CD4 microbeads | Miltenyi Biotec | 130-049-201 | |
| 2-Mercaptoethanol | Sigma | M7522 | |
| LS column | Miltenyi Biotec | 130-042-401 | |
| magnetic stand | Miltenyi Biotec | 130-042-303 | |
| 1ml syringe | Terumo | SS+01T | |
| Centrifuge | Eppendorf | Eppendorf 5810R | |
| Sony SY3200 cell sorter | Sony | Sony SY3200 cell sorter | |
| 50ml conical centrifuge tube | Greiner Bio-One | 210261 | |
| 15m conical centrifuge tube | Greiner Bio-One | 188271 | |
| FACS tube | Corning | 352054 | |
| 24 well cell culture plate | Greiner Bio-One | 662160 | |
| anti-mouse CD4 PerCP-eFluor 710 | eBioscience | 46-0041-82 | |
| PE conjugated anti-mouse CD25 (IL-2Ra, p55) | eBioscience | 12-0251-82 | |
| anti-human/mouse CD44 APC | eBioscience | 17-0441-82 | |
| anti-mouse CD62L FITC | eBioscience | 11-0621-85 | |
| Neubauer-improved counting chamber | Marienfeld | 640010 | |
| Hyclone Trypan Blue Solution | GE healthcare Life Sciences | SV30084.01 | |
| microscope | Nikon | Nikon Elipse TS100 | |
| Purified anti-mouse CD3e antibody | Biolegend | 100314 | |
| Purified hamster anti-mouse CD28 | BD Biosciences | 553295 | |
| Purified Rat anti-mouse IFNγ | eBioscience | 16-7312-85 | |
| Purified anti-mouse IL-4 Antibody | Biolegend | 504102 | |
| Recombinant Mouse IL-7 Protein | R&D system | 407-ML | |
| Recombinant Mouse IL-6 | R&D system | 406‑ML | |
| recombinant human TGF-beta | R&D system | 240-B-010 | |
| PMA | Merck Millipore | 19-144 | |
| Ionomycin | Sigma | I0634 | |
| GolgiPlug protein transport inhibitor | BD Biosciences | 51-2301KZ | |
| Intracellular Fixation&Permeabilization buffer set | eBioscience | 88-8824-00 | |
| anti-mouse GM-CSF PE | eBioscience | 12-7331-82 | |
| FITC anti-mouse IL-17A | Biolegend | 506908 | |
| APC anti-mouse IFN-gamma | Biolegend | 505810 | |
| GO Taq qPCR master mix | Promega | A6002 | |
| mouse GM-CSF ELISA Ready-SET-Go! | invitrogen | 88-7334-88 | |
| Mouse IL-17A Uncouted ELISA | invitrogen | 88-7371-22 |
Referências
- Zhu, J., Paul, W. E. CD4 T cells: fates, functions, and faults. Blood. 112, 1557-1569 (2008).
- Kara, E. E., et al. Tailored immune responses: novel effector helper T cell subsets in protective immunity. PLoS Pathogens. 10, e1003905 (2014).
- Bou Nasser Eddine, F., Ramia, E., Tosi, G., Forlani, G., Accolla, R. S. Tumor Immunology meets…Immunology: Modified cancer cells as professional APC for priming naive tumor-specific CD4+ T cells. Oncoimmunology. 6, e1356149 (2017).
- Dong, C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nature Reviews. Immunology. 8, 337-348 (2008).
- Korn, T., Bettelli, E., Oukka, M., Kuchroo, V. K. IL-17 and Th17 Cells. Annual Review of Immunology. 27, 485-517 (2009).
- Sheng, W., et al. STAT5 programs a distinct subset of GM-CSF-producing T helper cells that is essential for autoimmune neuroinflammation. Cell Research. 24, 1387-1402 (2014).
- Codarri, L., et al. RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nature Immunology. 12, 560-567 (2011).
- El-Behi, M., et al. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nature Immunology. 12, 568-575 (2011).
- Ivanov, I. I., et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126, 1121-1133 (2006).
- Zhang, J., et al. A novel subset of helper T cells promotes immune responses by secreting GM-CSF. Cell Death and Differentiation. 20, 1731-1741 (2013).
- Croxford, A. L., Spath, S., Becher, B. GM-CSF in Neuroinflammation: Licensing Myeloid Cells for Tissue Damage. Trends in Immunology. 36, 651-662 (2015).

