Using High Content Imaging to Quantify Target Engagement in Adherent Cells
Summary
Measurements of drug target engagement are central to effective drug development and chemical probe validation. Here, we detail a protocol for measuring drug-target engagement using high content imaging in a microplate-compatible adaption of the cellular thermal shift assay (CETSA).
Abstract
Quantitating the interaction of small molecules with their intended protein target is critical for drug development, target validation and chemical probe validation. Methods that measure this phenomenon without modification of the protein target or small molecule are particularly valuable though technically challenging. The cellular thermal shift assay (CETSA) is one technique to monitor target engagement in living cells. Here, we describe an adaptation of the original CETSA protocol, which allows for high throughput measurements while retaining subcellular localization at the single cell level. We believe this protocol offers important advances to the application of CETSA for in-depth characterization of compound-target interaction, especially in heterogeneous populations of cells.
Introduction
When developing new drugs or chemical probes it is essential to couple the observed pharmacological effect or functional readout to measurements of target occupancy or engagement in live cells1,2,3. These data are necessary both to ensure that the small molecule in fact reaches its desired target and to validate the biological hypothesis behind protein target selection4,5. Furthermore, during drug development, model systems of increasing complexity are used to select and corroborate a lead compound prior to clinical trials. To confirm translation of biology across these preclinical systems, methods for tracing drug-target engagement and accompanying biology throughout this development process are critical.
Drug-target engagement has traditionally been challenging to monitor in live cells with unfunctionalized small molecules and proteins, especially at the single-cell level with spatial resolution6,7. One recent method to observe the interaction between unmodified drugs and proteins in live cells is the cellular thermal shift assay (CETSA) in which ligand-induced stabilization of a native protein in response to a heat challenge is quantified8,9,10. This is accomplished by quantifying remaining soluble protein after exposure to a heat challenge. In the initial disclosure of CETSA, western blot was used for detection. To enable screening campaigns and hit triaging of larger compound collections, efforts to increase the throughput of CETSA experiments have lead to the development of several homogenous, microplate-based assays10,11. However, one limitation with these methods is that they are currently best suited to compound treatment in cell suspensions and the detection requires cell lysis, leading to loss of spatial information. CETSA can be applied experimentally either as a ligand-induced shift in thermal aggregation temperature (Tagg) at a single concentration of the small molecule or the ligand concentration necessary to stabilize the protein at a single temperature. The latter is termed isothermal dose response fingerprints (ITDRF) to signify the dependence of these measurements on the specific experimental conditions.
The goal of this protocol is to measure target engagement using CETSA in adherent cells by immunofluorescent (IF) antibody detection with high-content microscopy12. This procedure extends the original CETSA platform to allow for single-cell quantification of target engagement with conservation of subcellular localization. Notably, unlike many previous reports, in this procedure compound treatment is performed in live adherent cells without surface detachment or washing prior to the heat challenge, thus preserving the established binding equilibrium we aim to measure13. Currently, the method is validated for one target protein p38α (MAPK14) in several cell lines, and we hope that by sharing this procedure the technique can be applied broadly across the melting proteome. We anticipate that this protocol can be adapted throughout the drug development pipeline from screening, hit triaging through to monitoring of target engagement in vivo.
Protocol
1. Seeding of Cells
NOTE: For a general overview of the workflow see Figure 1. A detailed list of materials and reagents are available in the Table of Materials.
- Prior to seeding of the cells, drill holes with a standard drill in the frame of black 384-well imaging assay plates to avoid air bubbles being trapped under the plate later during the heating step. To avoid plastic particles entering the wells during this step and to maintain sterile conditions, seal the plates with an adhesive aluminum foil or cover the plate prior to drilling in a tissue culture hood. Typically, 3 holes with a diameter of 3.5 mm on each of side of the plate (short edge) suffice.
- Prepare a laminar flow bench by cleaning with 70% ethanol. Following standard aseptic tissue culture techniques, remove media from cell flask or dish. Wash cells with 5-10 mL of phosphate-buffered saline (PBS) and then add 2 mL trypsin to the flask. Incubate the flask at 37 °C until the A-431 cells detach. Count the cells either using a haemocytometer or cell counter. Prepare a cell suspension of 50,000 cells/mL in culture medium.
- Dispense 40 µL of cell suspension (giving a final cell density of 2,000 cells per well) into each well of an assay plate using a bulk reagent dispenser or a multichannel pipet, depending on the scale of the experiment. Briefly move the plate from side-to-side to disperse cells evenly on the bottom of the plate.
- To minimize plate-edge effects, allow the cells to settle at the bottom of the assay plate for 20 minutes at room temperature in the back of the laminar flow hood. Then, place the plate in a plastic container with damp paper towels to ensure a humid atmosphere. Prior to use, wipe the plastic container with 70% ethanol.
- Incubate the box with the assay plate for 2-3 days at 37 °C and 5% CO2 in a conventional humidified incubator. Monitor confluency of the cells until 50-75%, as assessed by visual inspection with a light microscope.
2. Compound Treatment
- On the day of the experiment, aspirate the medium from each well using a plate washer. Place the plate on the plate washer and select the aspiration program. If a plate washer is not available, then the liquid can be removed by inverting the plate with a rapid hand twist over a waste tray or sink. Complete removal of liquid is essential for good performance. Any excess liquid is then removed by dabbing with paper towels.
- Add 30 µL of compounds diluted to the appropriated concentration in cell culture medium using automated dispensing or a multichannel pipette depending on the scale of the experiment. Ensure to add a negative (DMSO) and positive (known ligand) control to several wells on each assay plate. Since this is a thermal shift assay, it is necessary that the compound concentration exceeds the dissociation constant to observe protein stabilization. Thus, a rough guideline for compound concentration is 50-100 times the IC50, but more detail descriptions for compound concentrations are found in the Discussion section.
NOTE: DMSO tolerability of the cell lines should be tested prior to the experiment. - Seal the compound-treated assay plate with a breathable plate seal and incubate at 37 °C and 5% CO2 in a humidified incubator for 30 minutes.
3. Heat Challenge
- First, set the water bath to the desired temperature. Note that the final temperature that is reached inside the wells of the assay plate can be different from the final temperature in the water bath. Investigate the offset beforehand with a dummy plate and thermocouple thermometer. It typically takes 30 minutes for the bath to stabilize at the desired temperature.
- To verify that the desired temperature is reached in the wells of the assay plate during the heating step, prepare an unsealed dummy plate containing the same volume of medium as the assay plate.
- Remove the assay plates from the incubator. Take off the breathable seal and re-seal the assay plate containing the compound-treated cells with a tight adhesive aluminum foil to ensure that no water will leak into the wells during the subsequent heating in the water bath. Ensure that the drilled holes in plate frame are accessible.
- Place the assay plate and the dummy plate in the water bath with the bottom of the plate angled towards the water surface to force any remaining air out from under the plates.
- Monitor the temperature inside the wells of a new dummy plate using a thermocouple thermometer.
- Heat the assay plate in the water bath for 3 minutes. Immediately transfer the assay and dummy plate to another water bath with room-tempered water to cool down for 5 minutes. The assay plate is now ready for further processing.
4. Fixation
- Dispense 10 µL 16% (w/v) paraformaldehyde (PFA) directly to the assay plate using a bulk reagent dispenser or a multichannel pipet. Incubate at room temperature for 20 minutes.
Note: Some fixatives are classified as carcinogenic and institutional safety regulations should be followed. - Aspirate the PFA solution and wash the cells with 300 µL PBS using a plate washer. Place the plate on the plate washer and select the aspiration program.
NOTE: This procedure has been optimized using an overflow protocol on the plate washer in which liquid is simultaneously dispensed and removed. If a plate washer or similar procedure is not available, the washing step can alternatively be done manually. - Manual washing procedure: Remove the liquid from the wells by inverting the plate with a rapid hand twist over a waste tray or sink. Complete removal of liquid is essential for good performance. Add 80 µL of PBS with a multichannel pipette and invert the plate again as described above, repeat the washing procedure two times. After the last wash, blot the plate against clean paper towels to remove any excess liquid.
5. Permeabilization
- Add 20 µL of 0.1% (v/v) NP-40 to the wells with a multichannel pipet and incubate at room temperature for 10 minutes. Wash the cells using the same procedure as described above (step 4.2).
- Alternatively, when appropriate, apply an antigen retrieval protocol, e.g.:
- Add 20 µL of 1% SDS to the wells with a multichannel pipet. Incubate at room temperature for 5 minutes and wash according to the same procedure described above (step 4.2).
- Add 80 μL of 10 mM glycine at pH 7.2 and incubate for 10 minutes at room temperature. Aspirate the glycine solution using a plate washer by placing on the plate washer and selecting the aspiration protocol. See notes in 2.1 and 4.2 if a plate washer is not available.
6. Blocking
- Add 15 µL of 1% (w/v) bovine serum albumin (BSA) in PBS to the wells using a bulk reagent dispenser or a multichannel pipet. Incubate the plate at room temperature for 1 hour or overnight at 4 °C with an aluminum foil plate seal.
7. Primary Antibody
- Aspirate the blocking solution using a plate washer by placing on the plate washer and selecting the aspiration protocol. See notes in 2.1 and 4.2 if a plate washer is not available.
- Add 10 µL of primary antibody diluted accordingly in 1% (w/v) BSA in PBS to the wells using a multichannel pipet. Incubate the plate at room temperature for 1 hour or overnight at 4 °C with an aluminum foil plate seal.
8. Secondary Antibody
- Aspirate the primary antibody solution and wash the wells according to same procedure as described above (step 4.2).
- Add 10 µL of Alexa 488 secondary antibody diluted accordingly in 1% (w/v) BSA in PBS. Incubate at room temperature for 1 hour. Seal the plate with an adhesive aluminum foil seal to protect from light.
NOTE: Protect the plate from light during this and subsequent steps.
9. Nuclear Staining and Cell Mask
- Add 10 µL of nuclear dye diluted to 0.05 mg/mL in PBS to the wells using a multichannel pipet. Incubate at room temperature for additional 10 minutes.
- Aspirate the secondary antibody and Hoechst solution and wash the wells using the same procedure as described above (step 4.2).
- Add 10 µL of cell mask diluted to 200 ng/mL in PBS to the wells using a multichannel pipet. Incubate at room temperature for 30 minutes.
- Aspirate the cell mask solution and wash the wells using the same procedure as described above (step 4.2).
- Dispense 60 µL of PBS to all wells using the plate washer, a bulk reagent dispenser, or a multichannel pipet, and seal the plates with an adhesive aluminum foil.
10. Image Acquisition and Analysis
- Capture images on a high content imager using 3 fluorescent channels: DAPI (387/447), GFP (472/520), and TexasRed (562/624). Acquire 4 images per well using 10X objective. Use automated laser autofocus and apply binning 2 during acquisition. Store images as 16 bit, gray scale tiff files along with metadata.
- Analyze images using available software. Identify cell boundaries using a Cell Scoring algorithm with DAPI (nucleus) and TexasRed (cytoplasm).
- Extract average intensity for all acquired wavelengths for further data analysis.
- Calculate the Z-factor to ensure the robustness of the assay.
- Calculate % stabilization using the following formula: 100*(1-(well intensity – average well intensity negative control)/(average intensity positive control-average intensity negative control)). Here negative control is DMSO and positive control is the reference substance. Since the maximum stabilization between compounds can vary and in fact be greater than the positive control for ITDRF curves, the maximum and minimum stabilization intensities for each compound are sometimes used in place of the intensity values of the control wells.
Representative Results
The protocol outlined in Figure 1 describes the basic workflow for running CETSA assays on adherent cells with detection of remaining soluble protein by high content imaging. This workflow can be easily adapted to all stages of assay development by modifying the plate layout of the compounds or reagents14. We detail expected results for several anticipated use cases below.
Antibody identification and assay development. A prerequisite for successful results is the identification of a primary antibody or other suitable affinity reagent that selectively recognizes the native form of the protein in the presence of the aggregated and precipitated protein formed during the heat challenge in step 3. To establish the CETSA imaging assay described here, we screened a panel of 9 antibodies targeting p38α at 52°C for signal window between the positive and negative controls. We then titrated the best antibodies and settled on the conditions shown in Figure 2 A,B with representative immunofluorescence images for p38α stabilized by a known ligand (positive control) and DMSO (negative control). Antibody recognition should also not be disrupted by the conformational changes of the target protein that may be induced by ligand binding (Figure 2C). As an example, BIRB796 has a long off rate, and quantification of target engagement was only possible by applying an antigen retrieval step (5.2; Figure 2D). It is important to validate the performance of the primary antibody with known ligands covering different binding sites of the target protein if available. The antibody validation is preferably done both with and without the antigen retrieval step.
Tagg and ITDRF curves. As mentioned above, CETSA experiments can be run in two different modes, Tagg curves and ITDRF experiments. Both variants utilize the same basic protocol outlined in Figure 1 and in the protocol section. In the first setup, the purpose is to challenge the cells with a temperature gradient and compare the Tagg curves in the presence and absence of a single concentration of ligand. To perform a Tagg curve, separate plates are heated for 3 minutes across a range of temperatures. In performing this experiment, it is important to time the compound treatment length for each plate with the time it takes for the water bath to stabilize to the new temperature. In this regard, peforming the heat challenge step in the water bath is more time consuming than heating tubes in a PCR machine. The experimental path is to next run concentration response curves of a ligand at a fixed temperature to generate ITDRF curves. In general, when testing multiple compounds, assay ready plates for the compound addition are prepared using automated liquid handling to achieve the most reproducible data. Compounds are serially diluted in DMSO and then dissolved in cell culture media to the desired concentrations. We have typically tested 11 point concentration series starting at 50 – 100 µM in 3 or 4 fold dilution, but this depends on the potency of the ligands used. It is advised to first establish the Tagg curve both in absence and presence of a ligand and select the temperature for subsequent isothermal experiments where a shift between the curves can be observed. The selected temperature should be around or just above the Tagg. Both formats allow for confirmation of target engagement but for ranking of compound affinities ITDRF experiments are often more suitable. Figure 3A shows an illustration of anticipated quantified results for a Tagg curve and Figure 3B quantifies results of a typical ITDRF experiment.
Screening campaign. The protocol can also be adapted to screening campaigns to identify novel binders of the target protein. In this case, isothermal heat challenges are applied for a large number of compounds at a single concentration followed by ITDRF experiments for identified stabilizing compounds. We prepare assay ready screening plates and transfer the compounds diluted in culture media to the assay plates using automated liquid handling. As with all thermal stability assays, it is necessary to exceed the dissociation constant to observe protein stabilization, and thus we have applied small molecule libraries at 50 µM to facilitate hit identification. Triaging of the hits using ITDRF will later allow ranking and prioritization of these compounds. Figure 4 shows a representative result from a screening plate.
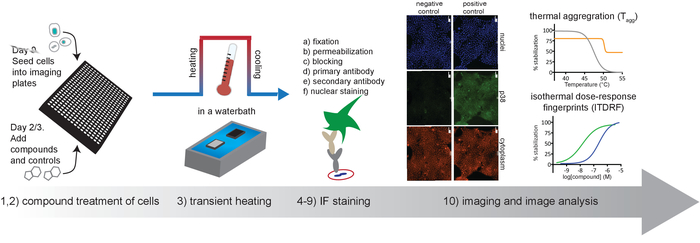
Figure 1. Schematic overview of the protocol described in this article. Please click here to view a larger version of this figure.
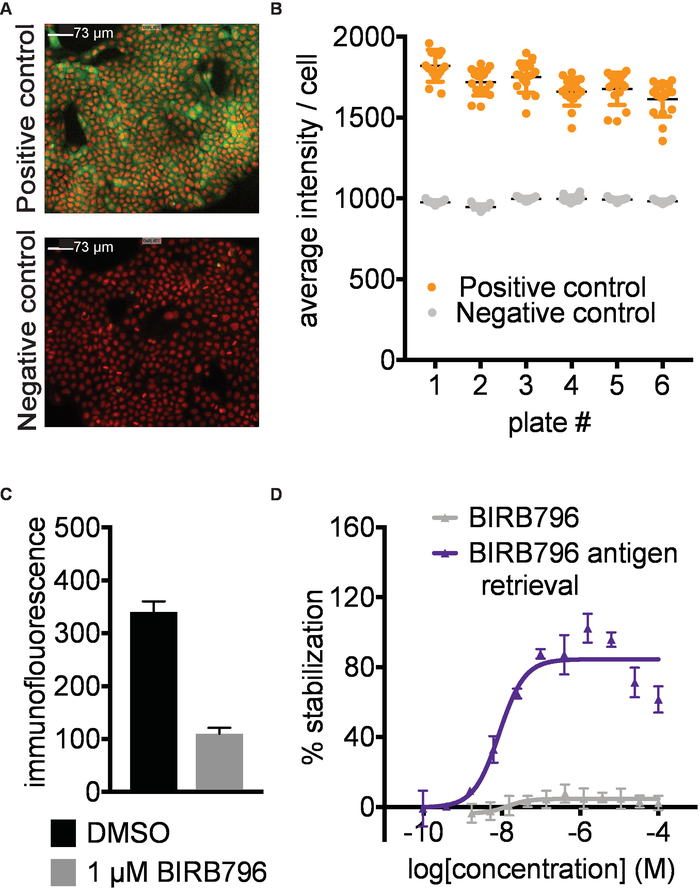
Figure 2. Antibody identification and assay development. A) Example data for detection of human p38α in A-431 cells. Representative images of positive (1 µM AMG548) and negative (DMSO) controls. Red- nuclear Hoechst staining, Green- p38α staining. Images taken with 10X magnification; the white scale bar represents 73 µm. B) Example of quantified data expressed as average intensity/cell. Error bars represent standard error of mean of 16 replicates for 6 separate plates. Each plate was heated to 52 °C in a water bath followed by fixation of the cells, permeabilization and subsequent immunostaining. C) Immunofluorescence signal intensity measured after treating A-431 cells with 1 µM BIRB796 or DMSO as described above followed directly by fixation. In the absence of a heating step, the BIRB796 signal is lower than the DMSO signal, suggesting that BIRB796 disrupts the detection of p38α with this antibody. D) ITDRFCETSA curves for cells treated with BIRB796 either with (blue triangle) or without (grey triangle) antigen retrieval protocol. This figure has been modified from Axelsson et al. 201812. Copyright 2018 American Chemical Society. Please click here to view a larger version of this figure.
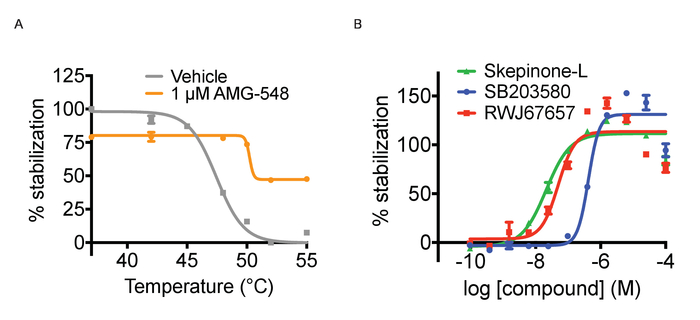
Figure 3. Thermal aggregation and ITDRF experiments. A) Thermal aggregation curve experiments for cells treated with positive (1 µM AMG548, in orange) and negative (DMSO in grey) controls. Error bars represent standard error of mean of 32 or 464 replicates. B) ITDRFCETSA experiment performed at 52 °C for cells treated with serial dilutions of SB203580 in blue, Skepinone-L in green and RWJ67657 in red. Error bars represent standard error of the mean of 6 replicates. Quantified data are expressed as average intensity/cell and normalized against the highest concentration of respective compound. This figure has been modified from Axelsson et al. 201812. Copyright 2018 American Chemical Society. Please click here to view a larger version of this figure.
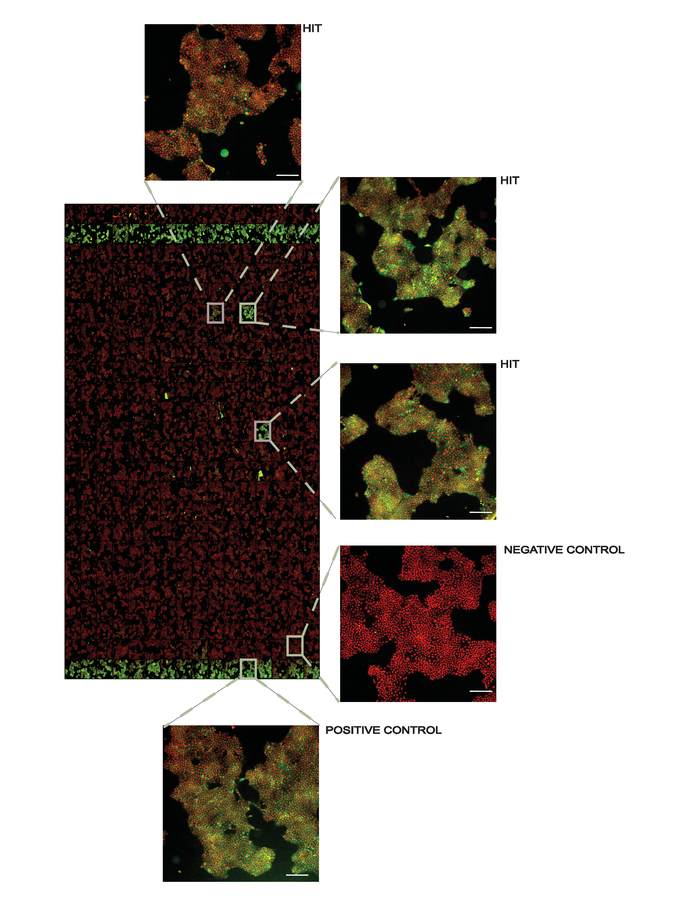
Figure 4. Representative overview of a screening plate at 10X magnification. Positive controls (1 µM AMG548) are in columns 2 and 24 and negative controls (DMSO) are in columns 1 and 23. All other wells contain 50 µM of the library compounds used for screening with several hits denoted. In the inset figures, the white line in the lower right corner represents 200 µm. This figure has been modified from Axelsson et al. 201812. Copyright 2018 American Chemical Society. Please click here to view a larger version of this figure.
Discussion
As discussed in the results section, there are several key steps to the procedure. First, it is important to identify a high-quality affinity reagent. We recommend screening a small library of antibodies for each desired target. After a primary antibody has been selected, it is also important to validate the system for a number of different binding sites of the protein target if appropriate. Counter-screening for compounds that interfere with the assay signal as shown in Figure 2C by omitting the heat challenge is highly encouraged. When a reduced signal in presence of a ligand is observed, an antigen retrieval protocol as described in step 5.2 can be tested. Uneven heating of the plates can cause fluctuations and plate-to-plate variations in the observed data. Since the plates are partly submerged in a water bath during the transient heat challenge (step 3.4) the outer wells of the plates are exposed to the hot water not only at the bottom of the wells but also through the outer wall. This may cause higher temperature during the same submersion time in the outer wells of the plate and subsequent plate edge effects. Therefore, it is recommended to avoid the outer wells of the plate. Similar effects can also be seen if air bubbles are trapped under the plate during the heating step preventing sufficient contact between the hot water and the bottom of the wells. Given the challenges with heating imaging plates, devices and plates specifically made for exposure to heat could help advance this method. We have explored the use of several cell lines, and have found variability in surface attachment after the heat challenge for adherent cell types. Preliminary experiments in our lab suggest that the use of a coating or extracellular matrix such as Synthemax II-SC or Cell-Tak can minimize cell detachment, but this could pose a technical challenge when using semi-attached cell types. Since compound treatment is performed in live cells, the small molecules must be cell permeable. If the compound is not membrane permeable, alternative high throughput CETSA methods are recommended. Some compounds need to be metabolically activated to bind to their target proteins11. In such cases, longer treatment cells with the compound prior to the heat challenge is recommended.
This protocol requires a single plate from cell seeding to imaging making it amenable to high throughput screening campaigns. The number of cells for each experiment is much lower than can be achieved with western blots or previous AlphaScreen assays15. Furthermore, washing or cell detachment steps, which may alter compound availability and binding, are removed, preserving the established binding equilibrium through the heat challenge step. Unlike previous procedures, a lack of signal due to cytotoxicity is simultaneously reported on by nuclear staining. Finally, imaging preserves the spatial resolution of the individual cells, allowing for target engagement measurements in mixed populations of cells or cell states.
To truly assess the applicability of the approach, concerted efforts to apply the technique across the melting proteome are needed. Such experiments will clarify the ease of identifying suitable affinity reagents, which report selectively on the native protein in the presence of the remaining denatured and aggregated proteins. We currently do not know how protein expression levels will affect the ability to reliably measure a signal window. We expect this to reflect affinity and selectivity of the available antibodies, and anticipate that for low abundant proteins technologies used to enhance signal in immunofluorescent assays will be applicable. Alternatively, tagged proteins or functionalized compounds could be used for detection in modified versions of the described protocol. This protocol is limited to targets that melt and for interactions where a quantifiable stabilization can be observed. For example, endogenous ligands can stabilize a protein in live cell assays, which may prevent further stabilization by an exogenous ligand. Although not explored here, we believe that it will be possible to multiplex readouts to allow for the study of multiple targets simultaneously, or a target with downstream effectors. Further studies are necessary to investigate these aspects of in situ CETSA.
Declarações
The authors have nothing to disclose.
Acknowledgements
The authors acknowledge infrastructure support from Science for Life Laboratory and Karolinska Institutet. The authors also acknowledge input and discussions with Michaela Vallin, Magdalena Otrocka and Thomas Lundbäck.
Materials
| Phosphate-buffered saline (PBS) | Medicago | 09-9400-100 | |
| TrypLE Express | ThermoFisher Scientific | 12604013 | for detaching cells and subculturing |
| 16% paraformaldehyde (PFA) | ThermoFisher Scientific | 28908 | fixative |
| Goat anti-rabbit IgG (H+L), Alexa Fluor 488 conjugated antibody | ThermoFisher Scientific | A11008 | secondary antibody |
| HCS CellMask Red stain | ThermoFisher Scientific | H32712 | Cytoplasm stain |
| NP-40 | Sigma-Aldrich | 56741 | for permeabilization |
| Hoechst stain 33342 | Sigma-Aldrich | B2261 | nuclear stain |
| Dulbecco’s modified Eagle’s medium (DMEM) – high Glucose | Sigma-Aldrich | 6429 | cell culture media component |
| Heat-inactivated fetal bovine serum (FBS) | Sigma-Aldrich | F9665 | cell culture media component |
| Penicillin-Streptomycin | Sigma-Aldrich | P4333 | cell culture media component |
| Corning, breathable plate seal | Sigma-Aldrich | CLS3345 | for copound incubation step |
| Rabbit anti-p38 antibody [E229] | Abcam | ab170099 | primary antibody, LOT:GR305364-16 |
| Falcon, Black 384-well clear bottom imaging plates | VWR | 736-2044 | imaging plates |
| Greiner, 384-well low volume polypropylene plates | VWR | 784201 | |
| Adhesive aluminum foil | VWR | 30127790 | |
| Peelable aluminium seal | Agilent | 24210-001 | for PlateLoc |
| LY2228820 | Selleckchem | S1494 | p38α inhibitor |
| PH797804 | Selleckchem | PH797804 | p38α inhibitor |
| BIRB796 | Selleckchem | S1574 | p38α inhibitor |
| SB203580 | Tocris | 1202 | p38α inhibitor |
| AMG 548 | Tocris | 3920 | p38α inhibitor |
| RWJ 67657 | Tocris | 2999 | p38α inhibitor |
| L-Skepinone | CBCS compound collection | p38α inhibitor | |
| Bovine serum albumin (BSA) | Sigma-Aldrich | A7030 | blocking agent |
| SDS (sodium dodecyl sulfate) | BDH | 44244 | used in antigen retrieval |
| Glycine | Sigma-Aldrich | G8898 | used in antigen retrieval |
| A-431 cells | ATCC | ATC-CRL-1555 | |
| Echo 550 | Labcyte | For preparation of compound plates | |
| Plate sealer | Agilent | PlateLoc | |
| Bulk reagent dispenser | Thermo Scientific | 5840300 | Multidrop Combi |
| Automated liquid handling | Agilent | Bravo liquid handling platform; used for compound plate preparation | |
| Plate washer | Tecan | Hydrospeed | |
| Water bath | Julabo | TW12 | |
| Thermocouple | VWR | Thermocouple traceable lab thermometer | |
| High content imager | Molecular Devices | ImageXpress Micro XLS Widefield High-Content Analysis System |
Referências
- Morgan, P., et al. Impact of a five-dimensional framework on R&D productivity at AstraZeneca. Nature Reviews Drug Discovery. 17 (3), 167-181 (2018).
- Freedman, L. P., Cockburn, I. M., Simcoe, T. S. The Economics of Reproducibility in Preclinical Research. PLOS Biology. 13 (6), e1002165 (2015).
- Waring, M. J., et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nature Reviews Drug Discovery. 14 (7), 475-486 (2015).
- Morgan, P., et al. Can the flow of medicines be improved? Fundamental pharmacokinetic and pharmacological principles toward improving Phase II survival. Drug Discovery Today. 17 (9), 419-424 (2012).
- Bunnage, M. E., Chekler, E. L. P., Jones, L. H. Target validation using chemical probes. Nature Chemical Biology. 9 (4), 195-199 (2013).
- Schürmann, M., Janning, P., Ziegler, S., Waldmann, H. Small-Molecule Target Engagement in Cells. Cell Chemical Biology. 23 (4), 435-441 (2016).
- Robers, M. B., et al. Target engagement and drug residence time can be observed in living cells with BRET. Nature Communications. 6, 10091 (2015).
- Martinez Molina, D., et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 341 (6141), 84-87 (2013).
- Martinez Molina, D., Nordlund, P. The Cellular Thermal Shift Assay: A Novel Biophysical Assay for In situ Drug Target Engagement and Mechanistic Biomarker Studies. Annual Review of Pharmacology and Toxicology. 56, 141-161 (2016).
- Jafari, R., et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nature Protocols. 9 (9), 2100-2122 (2014).
- Almqvist, H., et al. CETSA screening identifies known and novel thymidylate synthase inhibitors and slow intracellular activation of 5-fluorouracil. Nature Communications. 7, 11040 (2016).
- Axelsson, H., et al. In situ Target Engagement Studies in Adherent Cells. ACS Chemical Biology. 13 (4), 942-950 (2018).
- Seashore-Ludlow, B., Perspective Lundbäck, T. E. a. r. l. y. Microplate Application of the Cellular Thermal Shift Assay (CETSA). Journal of Biomolecular Screening. 21 (10), 1019-1033 (2016).
- Axelsson, H., Almqvist, H., Seashore-Ludlow, B., Lundback, T., Sittampalam, G. S., et al. . Assay Guidance Manual [Internet]. , (2016).
- Mateus, A., et al. Prediction of intracellular exposure bridges the gap between target- and cell-based drug discovery. Proceedings of the National Academy of Sciences of the United States of America. 114 (30), E6231-E6239 (2017).

