Quantifying the Heterogeneous Distribution of a Synaptic Protein in the Mouse Brain Using Immunofluorescence
Summary
Here, we describe a quantitative approach to determining the distribution of a synaptic protein relative to a marker protein using immunofluorescence staining, confocal microscopy, and computer-based analysis.
Abstract
The presence, absence, or levels of specific synaptic proteins can severely influence synaptic transmission. In addition to elucidating the function of a protein, it is vital to also determine its distribution. Here, we describe a protocol employing immunofluorescence, confocal microscopy, and computer-based analysis to determine the distribution of the synaptic protein Mover (also called TPRGL or SVAP30). We compare the distribution of Mover to that of the synaptic vesicle protein synaptophysin, thereby determining the distribution of Mover in a quantitative manner relative to the abundance of synaptic vesicles. Notably, this method could potentially be implemented to allow for comparison of the distribution of proteins using different antibodies or microscopes or across different studies. Our method circumvents the inherent variability of immunofluorescent stainings by yielding a ratio rather than absolute fluorescence levels. Additionally, the method we describe enables the researcher to analyze the distribution of a protein on different levels: from whole brain slices to brain regions to different subregions in one brain area, such as the different layers of the hippocampus or sensory cortices. Mover is a vertebrate-specific protein that is associated with synaptic vesicles. With this method, we show that Mover is heterogeneously distributed across brain areas, with high levels in the ventral pallidum, the septal nuclei, and the amygdala, and also within single brain areas, such as the different layers of the hippocampus.
Introduction
Communication between neurons happens at specialized contact sites called synapses. Synapses contain a myriad of different proteins that orchestrate synaptic transmission. Some of those proteins show a heterogeneous distribution throughout the nervous system and are not present in every synapse1. One example for such a protein is Munc13, which is involved in the priming process of synaptic vesicles. There are different isoforms of Munc13, which are heterogeneously distributed throughout the brain2, and the presence or absence of specific isoforms can influence short-term synaptic plasticity and synaptic vesicle dynamics3,4,5. Therefore, it is of vital importance to be able to identify the presence of different synaptic proteins across brain areas.
The methods of choice for quantification of synaptic proteins – so far – are mass spectrometry and Western blotting, rather than immunohistochemistry6,7,8,9. In some cases, several methods are used to complement each other to assess both the quantity and the localization of specific proteins (i.e., Wilhelm et al.10). The method we describe here allows for the localization and quantification of proteins of interest without the need of using any biochemical method, simply employing immunofluorescent stainings. Another advantage here is that the quantification can be done over areas much smaller and, therefore, more specific, than those achieved by other methods. However, one has to take into consideration that a reliable reference protein is needed to assess the distribution of the protein of interest.
Fluorescent staining by immunohistochemistry allows us to routinely identify the localization of proteins across brain areas as well as within different neuronal compartments. To identify the different compartments, specific markers are used. Typically, antibodies against synapsin and synaptophysin11 can be used to label synaptic vesicles, while antibodies against bassoon label the active zone of a presynaptic terminal12. Vesicular transporters, such as the vesicular glutamate transporters (vGluT) or vesicular GABA transporter (vGAT), are used to label excitatory13 and inhibitory14 presynaptic terminals, respectively. On the postsynaptic side, antibodies against the Homer protein can be employed to mark postsynaptic terminals, and antibodies against postsynaptic density protein 95 (PSD95)15,16,17 or gephyrin18,19,20 can label excitatory or inhibitory postsynaptic terminals, respectively. By using antibodies against a protein of interest and markers such as the ones described above, one can determine the localization of such protein. Many studies to date have done this in a qualitative manner21. However, to reliably determine the differential distribution of a specific synaptic protein, one must not only determine its presence or absence but also its relative concentration. The heterogeneity of sizes and density of synapses makes it important to establish a ratio between the synaptic marker and the protein of interest. Otherwise, synapse-rich regions such as the non-pyramidal layers of the hippocampus and the molecular layer of the cerebellum will show a high density of synaptic proteins, only due to the higher density of synapses but not due to a strong presence of that protein in each synapse (e.g., Wallrafen and Dresbach1). On the other hand, proteins in the neuronal soma (e.g., TGN3822) will usually show strong presence in the hippocampal pyramidal cell layer or hippocampal or cerebellar granule cell layer due to the high concentration of neuronal cell bodies in those areas. Therefore, this non-homogeneous distribution of structures, in this case synapses, can lead to a false estimation of the distribution of the protein of interest itself. Furthermore, there is an intrinsic variability in staining intensities across samples in immunohistochemical stainings. The protocol described here takes this into consideration and avoids such biases, as well as other caveats that arise from immunohistochemical methods.
In our recent study, we have used this method to describe the differential expression of Mover (also called TPRGL23 or SVAP3024) across 16 different brain areas1. Mover is a vertebrate-specific synaptic protein that can be found in association to synaptic vesicles and influences neurotransmitter release25,26,27. We have related the Mover expression to the abundance of synaptic vesicles, by staining for synaptophysin as a synaptic vesicle reference marker. We found high levels of Mover particularly in the septal nuclei, the ventral pallidum, and the amygdala. Within the hippocampus, we found a heterogeneous distribution of Mover, with high levels in the layers associated with intra-hippocampal computation, and low levels in input- and output layers.
Protocol
This protocol does not involve experiments on live animals. Experiments involving euthanizing of animals to obtain brain samples were approved by the local animal protection authorities (Tierschutzkommission der Universitätsmedizin Göttingen) under the approval number T 10/30.
NOTE: For this protocol, 3 adult male C57BL/6 mice were used.
1. Sample Preparation
- Prepare fixative and 0.1 M phosphate buffer (PB; see Table 1).
- Fix the animal by transcardial perfusion as described in Gage et al.28. First wash out the blood with 0.9% NaCl-solution, then perfuse with 30 mL of 4% paraformaldehyde (PFA).
- Open the skull with scissors and carefully isolate the brain using a spoon with blunt edges to avoid damaging the tissue.
- Fill a 50 mL reaction tube with fixative and postfix the brain in 4% PFA at 4 °C overnight.
- Remove the fixative and wash the brain in 50 mL of 0.1 M PB on a shaker for 30 min.
- After washing, incubate the brain in a 50 mL reaction tube in 30% sucrose in 0.1 M PB for 48 h or until it sinks in the tube at 4 °C for cryoprotection.
- Trim the cryoprotected brain with a sharp blade, place it in a cryomold, and embed it with optimal cutting temperature (OCT) compound. Avoid bubbles. Orient the brain and freeze the cryomold in the -80 °C freezer.
- Mount the frozen tissue for sectioning. Equilibrate the tissue to the cryomicrotome temperature for at least 15 min before sectioning.
- Section the brain into 25 µm thick coronal slices. Touch the OCT carefully with a glass hook without touching the brain tissue. Collect 3 adjacent slices per well in a 24 well plate and store them in 0.1 M PB at 4 °C until staining.
NOTE: The protocol can be paused here for up to two weeks. Longer storage times can interfere with the tissue quality and thus influence the outcome of the experiment.
2. Immunofluorescence
- Prepare solutions including the blocking buffer, antibody buffer, washing buffer 1, and washing buffer 2 (see Table 1).
- Rinse slices once with PB to remove excess OCT.
- Remove the solution with a plastic pipette without sucking in the brain slices. Add 250 µL of fresh PB with a 1000 µL pipette.
CAUTION: Slices should not dry out, so remove and add fluids well by well.
- Remove the solution with a plastic pipette without sucking in the brain slices. Add 250 µL of fresh PB with a 1000 µL pipette.
- Remove the PB with a plastic pipette and add 250 µL of blocking buffer per well with a 1000 µL pipette. Incubate for 3 h at room temperature (RT) on the shaker.
- During the incubation time, dilute the primary antibodies in antibody buffer in a reaction tube. Use 250 µL antibody buffer per well and add the appropriate amount of antibody (see Table 2) by pipetting it directly into the solution using a 2 µL pipette. Mix the solution by gently pipetting up and down several times. Vortex shortly afterwards to ensure proper mixing.
NOTE: To determine the background fluorescence, stainings should also be performed without adding the primary antibody. For that, incubate the slice in antibody solution without primary antibodies according to the protocol. - After the incubation time, remove the blocking buffer with a plastic pipette and add 250 µL of antibody solution containing primary antibodies per well. Incubate slices with primary antibody overnight at 4 °C on a shaker.
- Next day, wash the slices with washing buffer 1 3x for 10 min at RT on a shaker.
- Remove the medium with a plastic pipette and add 300 µL of washing buffer 1 per well. Incubate at RT for 10 min. Repeat 3 times.
- During the washing steps, dilute the fluorophore-coupled secondary antibodies in antibody buffer in a reaction tube. Use 250 µL antibody buffer per well and add the appropriate amount of antibody (see Table 2) by pipetting it directly into the solution using a 2 µL pipette. Mix the solution by gently pipetting up and down several times. Vortex shortly afterwards to ensure proper mixing.
CAUTION: Because the antibodies are light-sensitive, all steps from this point on need to be performed in the dark. - After the washing steps, remove the washing buffer with a plastic pipette and add 250 µL of antibody solution containing secondary antibodies per well. Incubate the slices with secondary antibody for 90 min at RT in the dark.
- Wash the slices with washing buffer 2 3x for 10 min at RT.
- During the washing steps, dilute 4′,6-diamidino-2-phenylindole (DAPI) in 0.1 M PB in a concentration of 1:2000.
- Remove the washing buffer 2 with a plastic pipette and add 250 µL of DAPI solution per well. Incubate for 5 min at RT on the shaker.
- Remove the DAPI solution with a plastic pipette and add 500 µL of 0.1 M PB per well with a 1000 µL pipette.
- Mount slices on microscope slides.
- Place a microscope slide under the stereoscope. With a fine brush, add three separate drops of 0.1 M PB onto the slide. Place one slice per drop onto the microscope slide.
- Use the fine brush to flatten and orient the slices on the microscope slide.
- When all slices are positioned correctly, remove excess PB with a tissue and dry the slide carefully.
CAUTION: Avoid drying the brain slices completely. - Add 80 µL of embedding medium onto the slide. Carefully lower the coverslip onto the slide, thereby embedding the brain slices.
- Leave the slides to dry in the fume hood for 1-2 h (cover them to avoid light exposure) and store them in a microscope slide box at 4 °C.
NOTE: The protocol can be paused here.
3. Imaging
- After the embedding medium is completely hardened, place the microscope slide under the confocal microscope.
NOTE: Epifluorescence microscopy combined with deconvolution software should yield similar image quality. - Adjust the laser settings by increasing or decreasing the laser intensity for every channel so that few pixels are overexposed to ensure maximum distribution of grey values.
- Acquire virtual tissues of the whole brain slice for the different channels.
- In the imaging software (see Table of Materials), select the Tiles option and manually delineate the brain slice with the Tile Region Setup.
- Distribute support points throughout the tile region and adjust the focus for the different support points by pressing Verify Tile Regions/Positions….
- Adjust the settings in Acquisition Mode according to the desired resolution and file size of the resulting image and start the scan.
- When the scan is finished, use the Stitching function to process the virtual tissue. Export the file as a .tif with the function Image Export.
4. Computer-based Analysis
- Load all single channels for one image into FIJI29 by clicking File| Open.
- With the Freehand selection tool, delineate one hemisphere in the DAPI-channel. Create a mask of the selection by clicking Edit| Selection| Create mask.
- Determine the mean fluorescence intensity for the single channels (Mover and synaptophysin) by clicking Analyze| Measure.
NOTE: Make sure to select the different channels to determine the mean fluorescence intensity values for each channel. - Copy the mean fluorescence intensity for the single channels into a spreadsheet.
- Determine the mean fluorescence intensity for the single channels in an area of interest by delineating the area also with the Freehand selection tool. Use a mouse brain atlas as reference.
- Repeat steps 4.1-4.5 for all hemispheres and all areas of interest.
NOTE: Determine the values for each hemisphere separately in order to later compare the values in an area of interest to that in the hemisphere (see step 5.2).
5. Data Handling
- In case the background fluorescence is high (see Discussion), a background subtraction might be needed. For that, determine the mean fluorescence intensity for the slice processed without primary antibody against the reference protein (here: synaptophysin) and subtract that value from all values obtained for the brain regions and hemispheres.
- When the mean fluorescence intensities for the single channels for every hemisphere and every area of interest have been determined (see Table 3), calculate the ratio of Mover to synaptophysin by dividing the value for Mover by the value for synaptophysin (yellow in Table 3). Perform this action for every hemisphere and every area of interest separately.
- Divide the ratio obtained for one area of interest by the ratio obtained for the corresponding hemisphere (orange in Table 3) to determine the ratio of the area of interest to the hemisphere.
- To determine the relative Mover abundance, translate the ratio obtained in 5.2 into a percentage by determining its deviation from 1 (red in Table 3). A ratio of 1.25 would therefore give a relative Mover abundance of 25% above average, and a ratio of 0.75 would yield a relative Mover abundance of 25% below average.
Representative Results
Representative staining patterns of different markers can be seen in Figure 1. The pattern varies depending on the distribution of the protein. Examples of five rostro-caudal levels are shown in columns (A)-(E). A representative DAPI staining is shown in the first row: DAPI adheres to the DNA of a cell and thus nuclei are stained. This results in a punctate pattern. Regions with a high cell density are brighter than regions with low cell densities. An example for a heterogeneously distributed protein can be seen in the second row. The Mover staining reveals a differential distribution throughout the brain, with bright hotspot areas and dimmer areas. In the third row, an example for the more homogeneously distributed reference marker synaptophysin is shown. An overlay of the two proteins (fourth row) shows the differential distribution of Mover (red) compared to the marker protein synaptophysin (green).
Figure 2 shows the quantification described in step 4 of the protocol. Shown are the mean fluorescence intensity values for the different channels across the hemispheres (Mover, Figure 2A; synaptophysin, Figure 2B) and across the areas of interest (Mover, Figure 2C; synaptophysin, Figure 2D). To determine the Mover abundance relative to the number of synaptic vesicles, a ratio is taken of the Mover fluorescence values to synaptophysin fluorescence values. These ratios for the areas of interest are shown in Figure 2E, and already provide an indication of the heterogeneous distribution of Mover, with areas with high and low Mover levels relative to synaptic vesicles. To additionally compensate for the inherent technical variability, the ratio in one area of interest (Figure 2E) is compared to that across the hemisphere (not shown) and translated into a percentage. This relative Mover abundance (Figure 2F) gives a measure of how much Mover is present in one area of interest relative to average.
As mentioned above, one of the major advantages of this technique is the ability to determine the abundance of the protein of interest across very small areas, even subregions and layers of areas of interest. One example of this application is shown in Figure 3, where the relative Mover abundance was determined for the different layers in the subfields of the hippocampus. The quantification in the different layers shown in Figure 3D, Figure 3F, and Figure 3H corresponds to the layers shown in Figure 3C, Figure 3E, and Figure 3G, with the corresponding colors. Within the hippocampus, Mover is heterogeneously distributed, with high Mover levels relative to synaptic vesicles in layers associated with intra-hippocampal computation (i.e., the polymorph layer of dentate gyrus [DG], stratum radiatum, lucidum and oriens of Cornu Ammonis 3 [CA3], and stratum radiatum and oriens of Cornu Ammonis 1 [CA1]), and low levels in input- and output layers (the inner and outer molecular layer of DG, the pyramidal cell layers of CA3 and CA1, and the stratum lacunosum-moleculare of CA1).
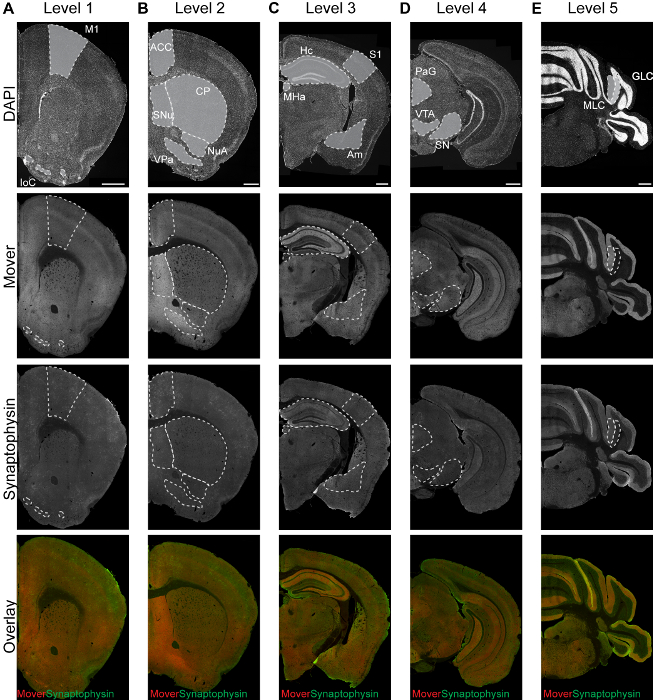
Figure 1: Representative immunofluorescence images of DAPI (first row), Mover (second row), synaptophysin (third row), and their overlay (fourth row, Mover in red, synaptophysin in green) at the 5 rostro-caudal levels (A-E). Areas of interest are shaded in grey in the upper row of panels. M1, primary motor cortex; IoC, islands of Calleja; ACC, anterior cingulate cortex; SNu, septal nuclei; VPa, ventral pallidum; NuA, nucleus accumbens; CP, caudate putamen; S1, primary somatosensory cortex; Hc, hippocampus; Am, amygdala; MHa, medial habenula; PAG, periaqueductal grey; SN, substantia nigra; VTA, ventral tegmental area; MLC, molecular layer of the cerebellum; GLC, granular layer of the cerebellum. Scale bar = 500 µm. This figure has been modified from Wallrafen and Dresbach1. Please click here to view a larger version of this figure.
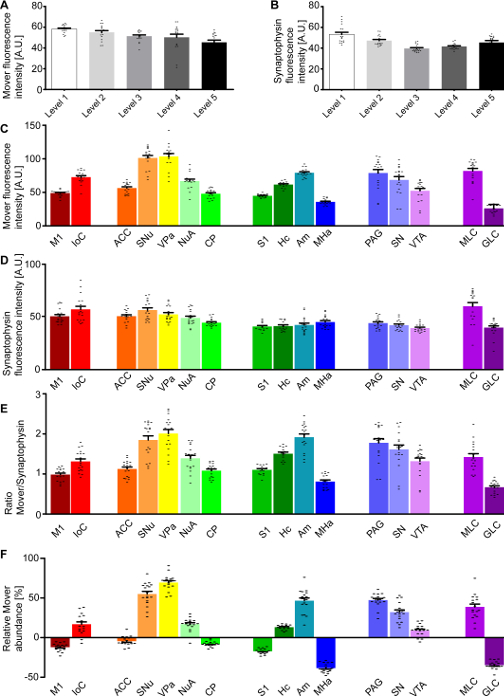
Figure 2: Quantification of the Mover distribution across the 5 rostro-caudal levels. Mean fluorescence intensity of the Mover signal (A) and the synaptophysin signal (B) at the different levels. Mean fluorescence intensity of the Mover signal (C) and the synaptophysin signal (D) at the 16 manually delineated brain regions. (E) Ratios of Mover and synaptophysin in the 16 brain areas of interest. (F) Quantification of the relative Mover abundance, comparing Mover/synaptophysin ratio at the respective region to the ratio of the corresponding hemisphere. M1, primary motor cortex; IoC, islands of Calleja; ACC, anterior cingulate cortex; SNu, septal nuclei; VPa, ventral pallidum; NuA, nucleus accumbens; CP, caudate putamen; S1, primary somatosensory cortex; Hc, hippocampus; Am, amygdala; MHa, medial habenula; PAG, periaqueductal grey; SN, substantia nigra; VTA, ventral tegmental area; MLC, molecular layer of the cerebellum. Black dots represent single data points. Bars show the mean ± standard error of the mean (SEM). This figure has been modified from Wallrafen and Dresbach1. Please click here to view a larger version of this figure.
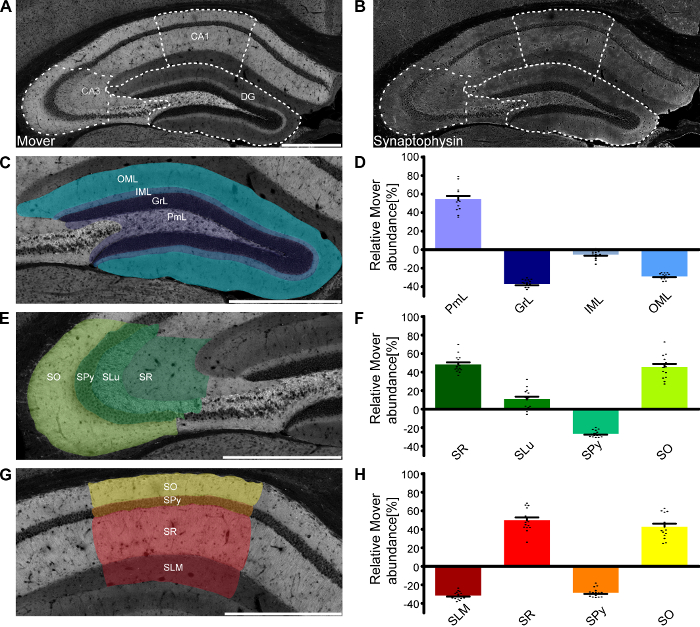
Figure 3: Mover distribution in the mouse hippocampus. Immunofluorescence stainings of coronal slices of the mouse hippocampus. Overview of the hippocampus showing the heterogeneous Mover expression pattern (A) and the corresponding Synaptophysin staining (B). The three regions of interest (DG, Figure 3C; CA3, Figure 3E; CA1, Figure 3G) are delineated with white dotted lines. (D,F,H) Quantification comparing the ratio in the respective layers to the ratio of the corresponding hemisphere. The colors in the bar graphs correspond to the respective shading in panels C, E, and G. Mover expression is high in levels associated with intra-hippocampal computation (i.e., the polymorph layer of DG, stratum radiatum, lucidum and oriens of CA3, and stratum radiatum and oriens of CA1), and low in the main input- and output layers (the inner and outer molecular layer of DG, the pyramidal cell layers of CA3 and CA1, and the stratum lacunosum-moleculare of CA1). OML, outer molecular layer; IML, inner molecular layer; GrL, granular layer; PmL, polymorph layer/hilus; SO, stratum oriens; SPy, stratum pyramidale; SLu, stratum lucidum; SR, stratum radiatum; SLM, stratum lacunosum-moleculare. Scale bar = 500 µm. Black dots represent single data points. Bars show the mean ± SEM. This figure has been modified from Wallrafen and Dresbach1. Please click here to view a larger version of this figure.
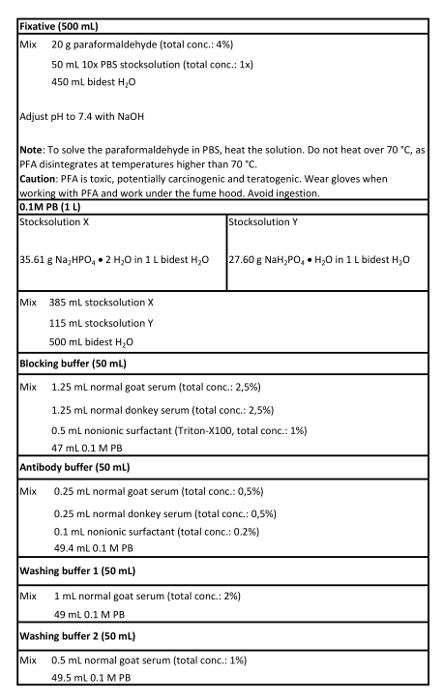
Table 1: Solutions used in this protocol.
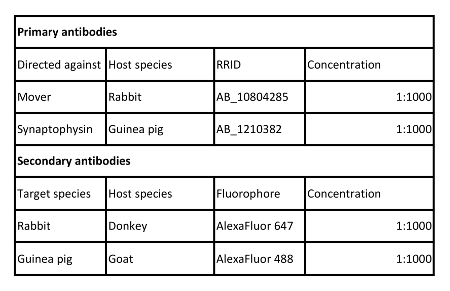
Table 2: Antibodies used in this protocol.
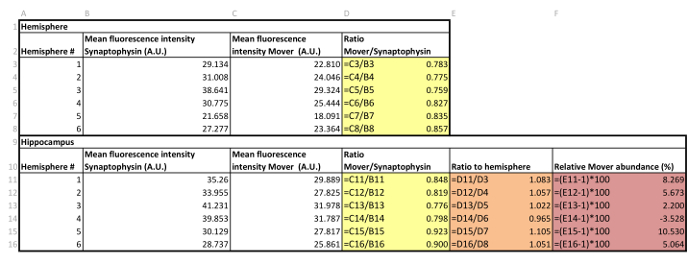
Table 3: Example of data handling.
Discussion
The method presented here aims at quantifying the distribution of a protein of interest relative to the abundance of a marker protein with a known distribution. Immunofluorescence staining can show a high variability of staining intensities between different slices. The quantification approach described here circumvents this problem by determining the ratio of the protein of interest to the average across the hemisphere. Therefore, different staining intensities across slices are cancelled out and allow for a quantitative description.
As with every immunofluorescence protocol, qualitative or quantitative, several factors can influence the success and thereby confound the analysis. Therefore, special attention should be paid to critical steps of the protocol. First, a proper fixation of the tissue is needed. This fixation can usually be achieved by a successful transcardial perfusion. The quality of perfusion can be verified by checking the liver shortly after washing out the blood. A first indicator for a successful perfusion is the clearing of the liver and extremities28. The presence of blood clots can indicate that the perfusion might have been too slow and should be performed faster next time. Some proteins require different fixation protocols, as chemical fixation with PFA can cause epitope blockage30. In this case, freeze fixation or fixation with a different chemical, such as methanol, should be considered. Second, after sectioning, it is critical to stain the brain slices as quickly as possible, preferably on the same or the next say. Longer storage in PB can lead to bacterial infection, and while adding NaN3 can prevent this to some extent, the tissue quality usually deteriorates with storage time. Third, during the staining procedure, it is important to perform washing steps well-by-well to avoid drying of the slices. When the slices dry out, background fluorescence can increase and thus cause a bias in the analysis. Fourth, after application of the secondary antibody, it is vital to perform all following steps in the dark. The fluorophores are light-sensitive, and light exposure can severely distort the fluorescence signal.
The optimization of the staining procedure, including the selection of adequate primary and secondary antibodies, optimal antibody concentration, and exposition time, is a prerequisite to achieve the best signal-to-noise ratio and to carry out a reliable quantitative immunofluorescence analysis. Antibodies verified in knock out tissue should be the preferred choice, albeit not always available. Always make sure to perform proper control experiments to exclude crosstalk between different antibodies. The amount of background fluorescence arising from autofluorescence and unspecific binding of the secondary antibody can be estimated by imaging the slices in which the primary antibody was not applied. It is not trivial to establish how much higher the intensities of the signal need to be when compared to the background to have an acceptable signal-to-noise ratio. However, based on empirical observations, the authors would suggest aiming for having a signal at least 2-fold stronger than background in order to reliably estimate the protein distribution. In case the background fluorescence is high in control conditions (without the presence of the primary antibody), the average background fluorescence should be subtracted from the experiment images.
The major advantage of our approach is its internal reference: the immunofluorescence intensity of the target protein (i.e., Mover) in a region of interest is compared to a reference marker (i.e., synaptophysin) and to the overall intensity of these proteins across the entire hemisphere. Thus, from the calculation we perform, one can unequivocally conclude that the abundance of the target protein is x fold higher/lower in a certain region of interest than the abundance of the reference protein relative to the distribution of the proteins across the entire hemisphere at this level relative to Bregma31. This allows for the comparison of results using different antibodies, different microscopes, or even across different studies. This consistency across different samples comes from the comparative nature of this method: variability is compensated for by taking the ratio between the fluorescence in the area of interest and that of the hemisphere. Therefore, dissimilarities in absolute values arising from technical differences are nullified. Another major advantage of this technique is the fact that the areas of interest can be as small as you want them to be, only limited by the resolution of the microscope. Quantifying protein levels with biochemical methods, for example Western Blot or mass spectrometry6,7,8,9, requires a dissection of the tissue into the area of interest. This dissection is hard for regions of the brain, such as the primary somatosensory cortex, and becomes virtually impossible when aiming for subregions, such as the different layers of the cortex or the hippocampus.
A caveat of the approach is that the different levels in the brain cannot directly be compared with each other. Hemispheres with many regions rich in the protein of interest will have a different average value than hemispheres with only few protein-rich regions. Values of 20% above average, for example, will therefore reflect a different absolute quantity of protein in one level relative to Bregma as compared to a second one. One has to keep in mind as well that this method does not allow the determination of absolute protein levels, only the relative abundance compared to the internal reference marker and the average across the hemisphere.
This method can be easily adapted to determine the distribution of the protein of interest relative to markers for different neuronal compartments, not only presynaptic sites. It can also be easily adapted for tissues other than the brain and – with suitable antibodies – other model systems than mice32,33. While the use of a confocal microscope is the authors' method of choice, a combination of epifluorescence microscopy and deconvolution software should yield the same data quality and thus expand the usability of the protocol. Additionally, the same stainings can be used to determine the subcellular distribution, for example with super-resolution microscopy.
Declarações
The authors have nothing to disclose.
Acknowledgements
We thank Irmgard Weiss for excellent technical assistance. The authors acknowledge support by Hermes Pofantis and Andoniya Petkova. The authors also thank the European Neuroscience Institute for the usage of the LSM800 and technical assistance, especially by Dr. Nils Halbsgut. This work was funded by the University Medical Center Göttingen. JSV acknowledges support by the Center for Nanoscale Microscopy and Molecular Physiology of the Brain (CNMPB).
Materials
| 1.5 mL reaction tubes | Eppendorf | 30120094 | |
| 50 mL reaction tubes | Greiner Bio-One | 227261 | |
| multiwell 24 well | Fisher Scientific | 087721H | |
| plastic pipette (disposable) | Sarstedt | 861,176 | |
| 1000 mL pipette | Rainin | 17014382 | |
| 2 ml pipette | Eppendorf | 3123000012 | |
| Vortex Genius 3 | IKA | 3340001 | |
| Menzel microscope slides | Fisher Scientific | 10144633CF | |
| Stereoscope | Leica | ||
| LSM800 | Zeiss | Confocal microscope | |
| freezing microtome | Leica | ||
| PBS (10X) | Roche | 11666789001 | |
| PFA | Sigma | P6148-1kg | |
| NaCl | BioFroxx | 1394KG001 | |
| sucrose | neoFroxx | 1104KG001 | |
| Tissue Tek | Sakura | 4583 | OCT |
| Na2HPO4 | BioFroxx | 5155KG001 | |
| NaH2PO4 | Merck | 1,063,460,500 | |
| normal goat serum | Merck Millipore | S26-100ML | |
| normal donkey serum | Merck | S30-100ML | |
| Triton X-100 | Merck | 1,086,031,000 | |
| rabbit anti-Mover | Synaptic Systems | RRID: AB_10804285 | |
| guinea pig anti-Synaptophysin | Synaptic Systems | RRID: AB_1210382 | |
| donkey anti-rabbit AF647 | Jackson ImmunoResearch | RRID: AB_2492288 | |
| goat anti-mouse AF488 | Jackson ImmunoResearch | RRID: AB_2337438 | |
| Mowiol4-88 | Calbiochem | 475904 | |
| ZEN2 blue software | Zeiss | Microscopy software | |
| FIJI | ImageJ | Analysis software | |
| Microsoft Excel | Microsoft | ||
Referências
- Wallrafen, R., Dresbach, T. The Presynaptic Protein Mover Is Differentially Expressed Across Brain Areas and Synapse Types. Frontiers in Neuroanatomy. 12, 58 (2018).
- Augustin, I., Betz, A., Herrmann, C., Jo, T., Brose, N. Differential expression of two novel Munc13 proteins in rat brain. Biochemical Journal. 337, 363-371 (1999).
- Rosenmund, C., et al. Differential Control of Vesicle Priming and Short-Term Plasticity by Munc13 Isoforms. Neuron. 33, 411-424 (2002).
- Breustedt, J., et al. Munc13-2 differentially affects hippocampal synaptic transmission and plasticity. Cerebral cortex. 20, 1109-1120 (2010).
- Chen, Z., Cooper, B., Kalla, S., Varoqueaux, F., Young, S. M. The Munc13 Proteins Differentially Regulate Readily Releasable Pool Dynamics and Calcium-Dependent Recovery at a Central Synapse. The Journal of Neuroscience. 33, 8336-8351 (2013).
- Taylor, S. C., Berkelman, T., Yadav, G., Hammond, M. A Defined Methodology for Reliable Quantification of Western Blot Data. Mol Biotechnol. , (2013).
- Charette, S. J., Lambert, H., Nadeau, P. J., Landry, J. Protein quantification by chemiluminescent Western blotting Elimination of the antibody factor by dilution series and calibration curve. Journal of Immunological Methods. 353, 148-150 (2010).
- Heidebrecht, F., Heidebrecht, A., Schulz, I., Behrens, S., Bader, A. Improved semiquantitative Western blot technique with increased quantification range. Journal of Immunological Methods. 345, 40-48 (2009).
- Toki, M. I., Cecchi, F., Hembrough, T., Syrigos, K. N., Rimm, D. L. Proof of the quantitative potential of immunofluorescence by mass spectrometry. Laboratory Investigation. 97, 329-334 (2017).
- Wilhelm, B. G., et al. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science. 344, 1023-1028 (2014).
- Navone, F., et al. Protein p38: An integral membrane protein specific for small vesicles of neurons and neuroendocrine cells. Journal of Cell Biology. 103, 2511-2527 (1986).
- Gundelfinger, E. D., Reissner, C., Garner, C. C. Role of Bassoon and Piccolo in Assembly and Molecular Organization of the Active Zone. Frontiers in Synaptic Neuroscience. 7, (2016).
- Ziegler, D. R., Cullinan, W. E., Herman, J. P. Distribution of vesicular glutamate transporter mRNA in rat hypothalamus. Journal of Comparative Neurology. 448, 217-229 (2002).
- Chaudhry, F. A., et al. The vesicular GABA transporter, VGAT, localizes to synaptic vesicles in sets of glycinergic as well as GABAergic neurons. Journal of Neuroscience. 18, 9733-9750 (1998).
- Aoki, C., et al. Electron Microscopic Immunocytochemical SAP-97 at Postsynaptic, Presynaptic, and Nonsynaptic Sites of Adult and Neonatal Rat Visual Cortex. Synapse. 257, 239-257 (2001).
- Valtschanoff, J. G., et al. Expression of NR2 Receptor Subunit in Rat Somatic Sensory Cortex : Synaptic Distribution and Colocalization With NR1 and PSD-95. Journal of Comparative Neurology. 611, 599-611 (1999).
- Hunt, A., Schenker, L. J., Kennedy, B. PSD-95 Is Associated with the Postsynaptic Density and Not with the Presynaptic Membrane at Forebrain Synapses. Journal of Neuroscience. 76, 1380-1388 (1996).
- Luscher, B., Fuchs, T., Kilpatrick, C. L. GABA A Receptor Trafficking-Mediated Plasticity of Inhibitory Synapses. Neuron. 70, 385-409 (2011).
- Kirby, M. . Understanding the molecular diversity of GABAergic synapses. 5, 1-12 (2011).
- Harris, K. M., Weinberg, R. J., Verrall, A. W. Ultrastructure of Synapses in the Mammalian Brain. Cold Spring Harbor Perspectives in Biology. , 1-30 (2012).
- Heise, C., et al. Selective Localization of Shanks to VGLUT1-Positive Excitatory Synapses in the Mouse Hippocampus. Frontiers in Cellular Neuroscience. 10, (2016).
- Peters, P. J., Yuan, L. C., Biology, C., Branch, M. Localization of TGN38 to the trans-Golgi network: involvement of a cytoplasmic tyrosine-containing sequence. Journal of Cell Biology. 120, 1123-1135 (1993).
- Antonini, D., et al. Tprg, a gene predominantly expressed in skin, is a direct target of the transcription factor p63. Journal of Investigative Dermatology. 128, 1676-1685 (2008).
- Burre, J., et al. Synaptic vesicle proteins under conditions of rest and activation: Analysis by 2-D difference gel electrophoresis. Electrophoresis. 27, 3488-3496 (2006).
- Kremer, T., et al. Mover is a novel vertebrate-specific presynaptic protein with differential distribution at subsets of CNS synapses. FEBS Letters. 581, 4727-4733 (2007).
- Ahmed, S., et al. Mover Is a Homomeric Phospho-Protein Present on Synaptic Vesicles. PLoS ONE. 8, (2013).
- Körber, C., et al. Modulation of Presynaptic Release Probability by the Vertebrate-Specific Protein Mover. Neuron. 87, 521-533 (2015).
- Gage, G. J., Kipke, D. R., Shain, W. Whole Animal Perfusion Fixation for Rodents. Journal of Visualized Experiments. , 1-9 (2012).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9, (2012).
- Schneider Gasser, E. M., et al. Immunofluorescence in brain sections: simultaneous detection of presynaptic and postsynaptic proteins in identified neurons. Nature Protocols. 1, 1887-1897 (2006).
- Paxinos, G., Franklin, K. B. J. . The Mouse Brain in Stereotaxic Coordinates. , (2001).
- Sun, Y., Ip, P., Chakrabartty, A. Simple Elimination of Background Fluorescence in Formalin-Fixed Human Brain Tissue for Immunofluorescence Microscopy. Journal of Visualized Experiments. , 1-6 (2017).
- Poole, A. R., Dingle, J. T., Mallia, A. K. The localization of retinol-binding protein in rat liver by immunofluorescence microscopy. Journal of Cell Science. 394, 379-394 (1975).

