A Robust Polymerase Chain Reaction-based Assay for Quantifying Cytosine-guanine-guanine Trinucleotide Repeats in Fragile X Mental Retardation-1 Gene
Summary
An accurate and robust polymerase chain reaction-based assay for quantifying cytosine-guanine-guanine trinucleotide repeats in the Fragile X mental retardation-1 gene facilitates molecular diagnosis and screening of Fragile X syndrome and Fragile X-related disorders with shorter turn-around time and investment in equipment.
Abstract
Fragile X syndrome (FXS) and associated disorders are caused by expansion of the cytosine-guanine-guanine (CGG) trinucleotide repeat in the 5’ untranslated region (UTR) of the Fragile X mental retardation-1 (FMR1) gene promoter. Conventionally, capillary electrophoresis fragment analysis on a genetic analyzer is used for the sizing of the CGG repeats of FMR1, but additional Southern blot analysis is required for exact measurement when the repeat number is higher than 200. Here, we present an accurate and robust polymerase chain reaction (PCR)-based method for quantification of the CGG repeats of FMR1. The first step of this test is PCR amplification of the repeat sequences in the 5’UTR of the FMR1 promoter using a Fragile X PCR kit, followed by purification of the PCR products and fragment sizing on a microfluidic capillary electrophoresis instrument, and subsequent interpretation of the number of CGG repeats by referencing standards with known repeats using the analysis software. This PCR-based assay is reproducible and capable of identifying the full range of CGG repeats of FMR1 promoters, including those with a repeat number of more than 200 (classified as full mutation), 55 to 200 (premutation), 46 to 54 (intermediate), and 10 to 45 (normal). It is a cost-effective method that facilitates classification of the FXS and Fragile X-associated disorders with robustness and rapid reporting time.
Introduction
Fragile X syndrome (FXS) and Fragile X associated disorders, e.g., tremor and ataxia syndrome (FX-TAS), and primary ovarian insufficiency (FX-POI) are mainly caused by cytosine-guanine-guanine (CGG) trinucleotide repeat expansion in the 5’ untranslated region (UTR) of the Fragile X mental retardation-1 (FMR1) gene on Xq27.31,2. The FMR1 encoded protein (FMRP) is a polyribosome-associated RNA-binding protein that functions in neuronal development and synaptic plasticity by regulating alternative splicing, stability, and dendritic transport of mRNA or modulating synthesis of partial postsynaptic proteins3,4,5,6,7.
The dynamic variation with a CGG repeat size of >200 is described as full mutation, which induces the aberrant hypermethylation and subsequent transcriptional silencing of the FMR1 promoter8. The resulting absence or lack of the FMRP protein disrupts normal neuronal development and causes FXS9, characterized by various clinical symptoms, including moderate to severe intellectual disability, developmental delay, hyperactive behaviors, poor contacts and autistic manifestations10,11,12. The presentation in female FXS patients is generally milder than that in males. The CGG repeat size ranging from 55 to 200 and 45 to 54 are classified as premutation and intermediate status, respectively. Due to the high degree of instability, the CGG repeat size in a premutation or intermediate allele presumably expands when transmitted from parents to offspring13,14. Thus, carriers with premutation alleles are at high risk of having children affected with FXS because of the repeat expansion, and in some cases, intermediate alleles can expand their repeat size to the full mutation range over two generations15,16. Furthermore, males with premutation also convey increased risk of developing late-onset FX-TAS17,18,19, while premutation females are predisposed for both FX-TAS and FX-POI20,21,22. Recently, it has been reported that autistic spectrum disorders with developmental delay and problems in social behaviors are presented in children with premutation FMR1 alleles23,24.
To determine the exact CGG repeat size is of great significance for classification and prediction of the FXS and Fragile X-associated disorders25,26. Historically, the CGG repeat region-specific polymerase chain reaction (PCR) with fragment sizing plus Southern blot analysis have been the gold standard for molecular profiling of the FMR1 CGG repeat27. However, traditional specific PCR is less sensitive to large premutations with more than 100 to 130 repeats and is incapable of amplifying full mutations27,28. Furthermore, capillary electrophoresis on a traditional genetic analyzer for repeat sizing fails to detect FMR1 PCR products with more than 200 CGG repeats. The Southern blot analysis enables differentiation of a wider range of repeat size, from normal to full mutation repeat numbers, and has been widely used for confirming full mutations (in males) and differentiating heterozygous alleles with a full mutation from apparently homozygous alleles with normal repeat sizes (in females). However, the resolution for quantifying the repeats is limited. More importantly, this step-by-step testing strategy is labor-intensive, time-consuming, and cost-ineffective.
Here, we present an accurate and robust PCR-based method for quantification of the CGG repeats of FMR1. The first step of this test is PCR amplification of the repeat sequences in the 5’UTR of the FMR1 promoter using Fragile X PCR kit. The PCR products are purified and fragment sizing is performed on a microfluidic capillary electrophoresis instrument, and subsequent interpretation of the number of CGG repeats using the analysis software by referencing standards with known repeats based on the rationale that PCR fragment length is directly proportional to the number of CGG repeats. The PCR system include reagents that facilitate the amplification of the highly GC-rich trinucleotide repeat region. This PCR-based assay is reproducible and capable of identifying all ranges of CGG repeats of FMR1 promoters. This is a cost-effective method that can find wide application in molecular diagnosis and screening of FXS and Fragile X-related disorders with less turn-around time and investment in equipment and thus, can be utilized in a broader spectrum of clinical laboratories.
Protocol
Ethical approval was granted by the Joint Chinese University of Hong Kong―New Territories East Cluster Clinical Research Ethics Committee (Reference Number: 2013.055)
1. PCR amplification
- Prior to starting, remove PCR buffer mix, sample diluent and DNA samples (both test and reference DNA) (see the Table of Materials) from the -20 °C freezer and keep them at room temperature for 20–30 min to make sure all reagents and DNA are fully thawed. Vortex and briefly spin down before use.
- Measure the concentration of the DNA samples using a spectrophotometer (see Table of Materials). The DNA concentration should be 25 ng/µL; dilute with sample diluent to the appropriate concentration if required.
NOTE: The DNA should be extracted and purified to remove interfering substances, such as proteins and high salt concentrations. Non-degraded, high-quality DNA should be used for subsequent PCR amplification and analysis (A260/A280: 1.8–2.0 and A260/A230: >1.0). - Label wells of a PCR plate or 0.2 mL PCR tubes to identify reference and tested DNA samples.
- Calculate the number of PCR reactions required for the test samples, 2 reference samples and negative control sample. Prepare PCR Master Mix by adding 15 µL of PCR buffer mix, 2.6 µL of sample diluent and 0.4 µL of polymerase for each reaction.
NOTE: A negative control using sample diluent is essential to monitor the PCR performance. Prepare the PCR master mix at room temperature, do NOT pipette on ice. The PCR buffer mix is viscous. Mix the tube and then briefly spin down prior to use. - Vortex the PCR master mix from step 1.4 for 10–20 s and spin down. Slowly dispense 18 µL of the mixture into each well or tube.
- Vortex and spin down the DNA samples. Pipette 2 µL of each DNA into the appropriate well or tube for a final PCR volume of 20 µL. Mix by pipetting up and down 5 times.
NOTE: The total amount of DNA per reaction should be 50–100 ng. DNA amounts greater than 150 ng per 20 µL PCR reaction may result in a poor amplification of large repeat alleles. For lower concentrated DNAs, the amount of sample diluent in the PCR master mix can be replaced by DNA solution. - Seal the plate with adhesive plate sealer, or with tube caps.
- Place the sealed PCR plate or tubes in the thermal cycler with heated lid. Run the program with the following settings: 95 °C for 5 min, followed by 25 cycles of denaturing at 98 °C for 35 s, annealing at 59 °C for 35 s, and extension at 72 °C for 4 min; a final step at 72 °C for 10 min. Hold the PCR products at 4 °C in the cycler until removal for further processing.
- After PCR amplification, purify and analyze the products immediately, or store at +2 to +8 °C overnight. Alternatively, the product can be stored for up to 30 days at -30 to -16 °C.
2. Purification of the PCR Products
- Preheat the incubator shaker to 65 °C.
- For each PCR reaction, add 80 µL of 1x TE buffer (see the Table of Materials) to the 20 µL of each PCR product from section 1.
- Transfer the sample mixture into a PCR clean-up plate (see Table of Materials) using a multichannel pipette.
- Keep the plate uncovered and place it into the incubator shaker, and incubate at 65 °C while shaking at 1,200 rpm for 10 min.
- After incubation, set the vacuum instrument at 250 mbar (or 25 kPa, 188 mmHg, 7.4 in Hg) and aspirate the solution through the filter for 15 min. Wells should have no liquid remaining.
- Cool the incubator shaker down to 25 °C.
- After the first aspiration, turn off the vacuum and add 50 µL of 1x TE buffer to each well. Do not mix. Aspirate the solution for 10 min using the vacuum settings in step 2.5.
- Dry the bottom of the filter plate by pressing it firmly on a stack of paper towels.
- Add 20 µL of 1x TE buffer into the bottom center of each well. Place the plate in the incubator shaker and incubate at 25 °C while shaking at 1,200 rpm for 5 min.
- After incubation, transfer >15 µL of each purified PCR DNA from step 2.9 to a fresh 96-well PCR plate. The purified DNA can be analyzed directly, or alternatively can be stored at -30 to -16 °C until required.
3. Fragment Sizing of PCR Products
- Prior to starting, allow DNA dye concentrate, DNA gel matrix, DNA marker, DNA ladder and purified DNA samples from step 2 to equilibrate to room temperature for 30 min.
- Set up the priming station.
- Replace the syringe (see Table of Materials) when using a new batch of reagents.
- Adjust the base plate, and release the lever of the syringe clip and slide it up to the top position.
- Start the sizing software (see Table of Materials) and prepare the gel-dye mix.
- Vortex dye concentrate for 10 s and spin down. Add 25 μL of the dye to a gel matrix vial. Vortex the mixed solution well and spin down.
- Transfer the gel-dye mix to a spin filter. Place the spin filter in a microcentrifuge and spin for 10 min at room temperature at 1,500 x g ± 20%.
NOTE: Protect the solution with dye from light and store at 4 °C after use. The gel-dye mix can be used for about 15 chips once prepared. Allow the gel-dye mix to equilibrate to room temperature for 30 min each time before use.
- Transfer the gel-dye mix to a spin filter. Place the spin filter in a microcentrifuge and spin for 10 min at room temperature at 1,500 x g ± 20%.
- Load the gel-dye mix.
- Insert a new DNA chip on the priming station. Add 9 μL of gel-dye mix into the well marked with “G”. Please ensure the plunger is positioned at the 1 mL mark and then close the priming station.
- Press the syringe plunger down until it is held by the clip. Wait for exactly 30 s then release the clip. Wait for 5 s, and then slowly pull the plunger back to the 1 mL position.
- Open the priming station and add 9 μL of gel-dye mix into the wells marked with “G”.
- Add 5 μL of marker into the well marked with the ladder symbol and also add 5 μL of marker into each of the 12 sample wells. Do not leave any wells empty.
- Add 1 µL of DNA ladder into the well marked with the ladder symbol. Add 1 µL of PCR product (used wells) from step 3.1 or 1 µL of ultrapure water (unused wells) into each of the 12 sample wells. Put the chip horizontally in the adapter of vortex mixer and vortex for 1 min at the indicated setting (2,400 rpm).
- Insert the chip in the bioanalyzer instrument and run the chip in the instrument within 5 min.
- After the assay is complete, immediately remove the used chip from the instrument.
- Slowly add 350 μL of deionized water into one of the wells of the electrode cleaner. Open the lid of the bioanalyzer and place the electrode cleaner into it. Close the lid and incubate for about 10 s. Open the lid and remove the electrode cleaner. Wait another 10 s to allow the water on the electrodes to evaporate and then close the lid.
4. Analyze the Fragment Sizing Results
NOTE: The reference samples should be amplified and analyzed by the same thermal cycler and bioanalyzer in the same batch with the unknown samples.
- After the bioanalyzer run completes, export the peak data from each run as a .csv table file for subsequent analysis.
- Start the analysis software and open the exported .csv peak table file from step 4.1.
- Through the QC menu tab, review the regression line fitted to the four points (shown as blue diamonds on the plot) from the two reference samples. The R2 value of the regression line should be >0.98 (typical values exceed 0.999).
- Through the Resultados menu tab, check the repeat size of each sample whose fragment length(s) is automatically plotted against the linear regression standard curve derived from the reference samples. The software also provides the classification of each sample according to different guidelines.
- Through the Export menu tab, export the result report for each sample with the repeat numbers and diagnostic classification, as well as a summary of sample information and QC report for each run.
NOTE: Analysis software allows the use of custom classification guidelines, such as American College of Medical Genetics (ACMG) or Clinical Molecular Genetics Society/ European Society of Human Genetics (CMGS/ESHG) guidelines, as well as predefined classification criteria.
Representative Results
The sizing results of the premutation female reference sample (NA20240, repeat sizes of 30 and 80) and the full mutation female reference sample (NA20239, repeat sizes of 20 and 200) are shown in Figure 1A and Figure 1B, respectively. Typically, two marker peaks (lower marker 50 base pairs [bp] and upper marker 10,380 bp) are included in the fragment size profile. There is usually a primer complex peak with a size of nearly 95 bp. Through the reference sample, a linear regression standard curve with four points can be constructed, as exhibited in Figure 1C.
The representative size features of clinical normal, intermediate, premutation, and full mutation samples are shown in Figure 2A–D. Particularly, mosaic full mutations with two expanded fragment peaks and one normal peak are presented in Figure 2E. In some female cases, only one peak is displayed in the microfluidic electrophoresis results, as shown in Figure 2F. This can be explained by the presentation of normal homozygous alleles (with the same CGG repeat numbers in the two alleles) in these cases or the inability of differentiating heterozygous alleles that have repeat number differences of four or less29. However, these single-peak results could be classified as normal, because it has been validated that the PCR-based pipeline enables robust amplification and detection of full mutation or premutation alleles, which minimizes the possibility of false negative in this situation. One type of sub-optimal situation is baseline bias, as shown in Figure 2G, which could produce ambiguous or uninterpretable results in some cases. Such a condition is suspected to be caused by an instrument issue. The measured fragment sizes of unknown samples are plotted against the linear regression standard curve derived from the reference samples to automatically calculate the repeat sizes using the analysis software, as displayed in Figure 2H. Fragment sizes less than 200 repeats are interpolated into the linear regression standard curve while larger full-mutation allele sizes are measured by extrapolating along the same standard line. The software also displays the classification of each sample according to different guidelines.
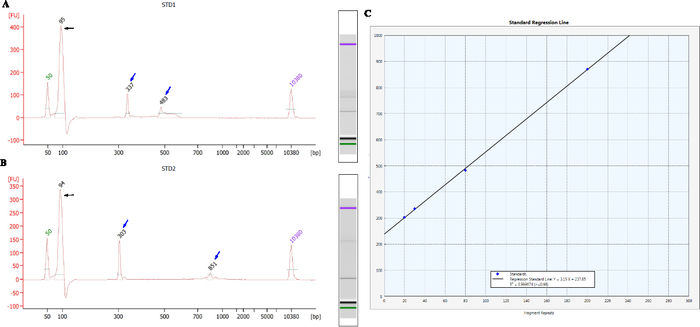
Figure 1: The sizing results of the female reference samples and the corresponding linear regression curve. (A, B) show the size features of the premutation female reference sample (NA20240, repeat sizes of 30 and 80) and the full mutation female reference sample (NA20239, repeat sizes of 20 and 200), respectively. Two markers (lower marker, 50 bp and upper marker 10380 bp) are included in the electrophoresis result for each sample. The peak with a size of nearly 95 bp indicates a primer complex. Black arrows indicate the peak size of primer complex, and blue arrows represent the fragment length of reference sample. (C) The linear regression standard curve with four points (blue diamonds) from the two reference samples is constructed in the analysis software. The horizontal and vertical axes show the calculated CGG repeat numbers and the measured fragment length in microfluidic electrophoresis. The R2 value of the regression was 0.99967. Please click here to view a larger version of this figure.
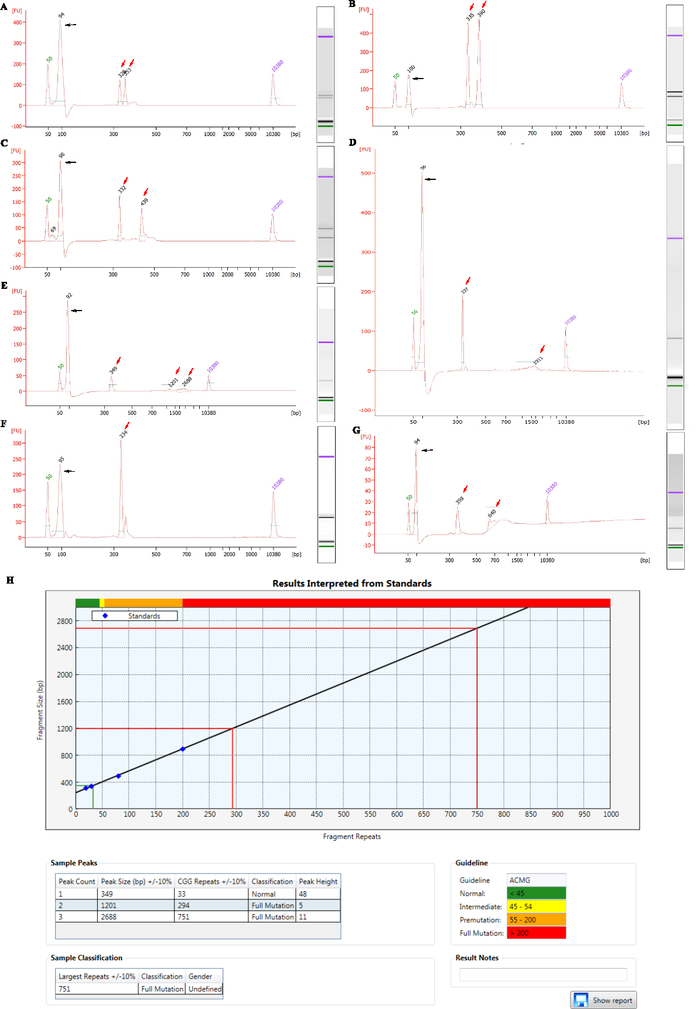
Figure 2: The representative results of clinical samples with different CGG repeat size. (A–E) show the representative size features by bioanalyzer in the order of normal (with fragment lengths of 328 bp and 353 bp, corresponding to the repeat numbers of 28 and 36), intermediate (with fragment lengths of 335 bp and 390 bp, corresponding to the repeat numbers of 30 and 49), premutation (with fragment lengths of 332 bp and 439 bp, corresponding to the repeat numbers of 29 and 65), full mutation (with fragment lengths of 337 bp and 1911bp, corresponding to the repeat numbers of 31 and 545) and mosaic full mutations (with fragment lengths of 349 bp, 1201 bp and 2688 bp, corresponding to the repeat numbers of 33, 294 and 751) samples. (F) presents a single-peak microfluidic electrophoresis result (with a fragment length of 334 bp, corresponding to the repeat number of 30) of a female who probably has homozygous alleles (with the same CGG repeat numbers in the two alleles) or heterozygous alleles (with the CGG repeat number differences of four or less). (G) shows one type of the sub-optimal situation that is baseline bias. Black arrows indicate the peak size of primer complex, and red arrows represent the fragment length of the sample. (H) displays the main result interface of the analysis software. The measured fragment sizes of unknown samples are plotted against the linear regression standard curve to automatically calculate the repeat sizes. The normal allele of the mosaic full-mutation female sample shown in (E) is mapped to the standard curve at the lower left corner of the coordinate axis region (plotted in green), and the larger full-mutation alleles (plotted in red) extrapolated beyond the four standard points (blue diamonds on the curve). The tabular sections in the lower half of the figure present the fragment sizes and the corresponding diagnostic classification according to the chosen ACMG guideline boundaries. Please click here to view a larger version of this figure.
Discussion
FXS is the second most common cause of intellectual impairment after trisomy 21, accounting for nearly one-half of X-linked mental retardation30, which may affect approximately 1 in 4,000 males and 1 in 8,000 females. More importantly, nearly 1 in 250–1,000 females carry a premutation, and this frequency is 1 in 250–1,600 in males26,31,32,33. Since the risk of CGG repeat expansion to full mutations when transmitting premutation alleles to offspring dramatically elevates, for example, from 4% when the maternal repeat size is 55–59 to 98% when the size is 100–20014, the determination of the CGG repeat size at a wider range could facilitate diagnosis of FXS and screening of premutation carriers for their reproductive planning. The PCR-based method introduced here is accurate, rapid and robust to amplify the repeat sequences in the 5’UTR of the FMR1 promoter and quantify the full spectrum of CGG repeat numbers on a microfluidic capillary electrophoresis instrument, and thus can boost its wide application in molecular diagnosis and screening of FXS and Fragile X-related disorders with less turn-around time and investment in equipment. The bioanalyzer instrument can be confidently utilized in low to moderate throughput test settings for repeat size measurement. The equipment is much smaller, less costly and simpler to maintain than other capillary electrophoresis instruments, such as ABI capillary genetic analyzers. For high volume screening, the MultiDX system containing up to 384-sample model provides an extensive flexibility and throughput for the tests.
The assay includes four main steps: PCR amplification of the repeat sequences in the 5’UTR of the FMR1 promoter (PCR set-up and PCR amplification takes nearly 3.5 h), purification of the PCR products (takes nearly 1 h), fragment sizing on a microfluidic capillary electrophoresis instrument (takes nearly 1 h), and interpretation of the number of CGG repeats using the analysis software by inferring from the reference standards with known repeats (takes nearly 0.5 h). In total, the turn-around time of this assay is approximately 6 h. For optimal performance, DNA samples should be purified to remove putative interfering substances such as proteins and high salt concentrations. Additionally, the recommended amount of input DNA is 50–100 ng per 20 µL of PCR reaction. DNA amounts greater than 150 ng per 20 µL of the PCR system have been shown to result in poor amplification of large repeat alleles. Furthermore, it is strongly recommended to set up at least two reference samples with well characterized repeat sizes in each PCR run for simultaneous analysis of repeat numbers as quality control and subsequent automated fragment sizing in the analysis software29. Typically, the R2 value of the linear regression curve derived from the reference samples should be greater than 0.98. In addition, the PCR products should be purified before microfluidic capillary electrophoresis to improve detection efficiency. As the PCR primer is labeled with FAM fluorescence29, fragment sizing with an appropriate electrophoresis system (e.g., bioanalyzer and the MultiDX system) can be directly performed without additional labeling.
A variety of methodologies for the diagnosis and study of FXS and related disorders have been comprehensively reviewed by Bruce E. Hayward et al., including DNA-based assays and FMRP protein assays34. As in most cases the molecular basis of FXS is dynamic mutation characterized by an expansion CGG repeat in the promoter region of FMRP1 gene, Southern blotting and amplification-based assays by using genomic DNA are the most commonly used assays for determination of the repeat number. It is well known that PCR-based assays overweigh Southern blotting in cost-effectiveness and minimum DNA amount requirement. However, amplification of the CGG-repeat is a main challenge since high GC content can affect the efficiency, and much effort has been made for the optimization of the PCR system over the years34. The fragile X PCR kit has been optimized for accurate amplification of the entire CGG trinucleotide repeats, and a validation study on the performance of this PCR-based method has been previously reported by our group29. A similar PCR-based method was reported by Mailick Seltzer et al. in 201235, but the ABI 3730xl instrument was used for repeat size determination and only individuals with repeat size less than 200 were identified. This may be due to the fact that their chosen platform was unable to detect the larger repeats with repeat size above 200. In contrast, the bioanalyzer instrument we use offers a more robust fragment sizing assay, as it can accurately and cost-effectively detect the full spectrum of the FMR1 CGG repeat including full mutations29.
Except repeat number, several other factors also need to be ascertained for appropriate diagnosis of FXS and FXS-related disorders, including FMR1 mutation, the extent of methylation and mosaicism34. The methylation status can be monitored by various methods such as incorporation of pre-digestion steps using methylation-sensitive restriction enzyme or bisulfite modification assay34, however, our assay is incapable of determining the methylation status of the 5’UTR of the FMR1 promoter. Additionally, the expansion risk of a FMR1 allele also depends on the presence of AGG interruptions, which can stabilize the gene during transmission. PCR-based repeat assays have been modified to detect AGG-interruptions whereas digestion enzyme instability is one of the main limitations. Triplet-primed (TP) PCR by using a hybrid PCR primer binding in CGG repeats or AGG interruptions is another type of PCR assay which can detect CGG repeat size and AGG interruptions simultaneously. However, the FMR1 gene-specific PCR method we have described is unable to identify the AGG interruptions unless FMR1 gene sequencing after the amplification is performed. Recently, Hayward et al. reported the PCR methods used in their own laboratory for determination all the parameters necessary for a complete genetic workup or thorough laboratory study, including assays that can detect repeat number, AGG interruption status and methylation status36. Unfortunately, neither assay is capable of comprehensively determining all these factors up to date.
It is worth noting that the assay is unable to detect deletions or single-nucleotide variants within the FMR1 gene, which accounts for approximately 1% of the FXS cases27. Rarely, in individuals who have cellular mosaicism for the FMR1 repeat, PCR may give a false negative result because of failure in detecting mosaicism for larger premutation and full-mutation alleles37,38. It has been demonstrated that the threshold of this PCR-based assay for mosaic full mutation male sample (341 repeats) is 2.5% when the peak detector threshold is set at three fluorescence units above baseline29. Southern blot analysis is recommended in the cases if mosaic alleles are indicated. Furthermore, with regards to the electrophoresis platforms, previous study has indicated that compared with capillary electrophoresis systems (e.g., ABI 3130XL instrument), the bioanalyzer instrument is incapable of differentiating the normal homozygous female samples with individual fragment peaks from female samples with heterozygous repeat sizes that have repeat number differences of four or less29. However, these single-peak samples could be classified as normal, because it has been validated that this PCR-based pipeline enables robust amplification and detection of full mutation or premutation alleles, which minimizes the possibility of false negative in this situation. Regardless of the above limitations, the method can be utilized as a first-tier pipeline for molecular identification of FXS and Fragile X-associated diseases with cost-effectiveness, robustness and rapid reporting time, supplemented by sequencing of the FMR1 gene and Southern blot analysis of methylation levels.
Declarações
The authors have nothing to disclose.
Acknowledgements
This research was supported by grants from NSFC Emergency Management Project (Grant No. 81741004), the National Natural Science Foundation of China (Grant No. 81860272), the Major Research Plan of the Provincial Science and Technology Foundation of Guangxi (Grant No. AB16380219), the China Postdoctoral Science Foundation Grant (Grant No. 2018M630993), and the Guangxi Natural Science Foundation (Grant No. 2018GXNSFAA281067).
Materials
| Agilent 2100 Bioanalyzer instrument: 0.2 mL PCR tubes | Axygen | PCR-02D-C | |
| Agilent 2100 Bioanalyzer instrument: 1X TE buffer, pH 8.0, Rnase-free | Ambion | AM9849 | |
| Agilent 2100 Bioanalyzer instrument: 2100 Bioanalyzer instrument | Agilent | G2939AA | |
| Agilent 2100 Bioanalyzer instrument: 96-well PCR Plate | Thermo Fisher | AB0800 | |
| Agilent 2100 Bioanalyzer instrument: Electrode cartridge | Agilent | Supplies equipment of the 2100 Bioanayzer instrument | |
| Agilent 2100 Bioanalyzer instrument: IKA vortex mixer | Agilent | Supplies equipment of the 2100 Bioanayzer instrument | |
| Agilent 2100 Bioanalyzer instrument: Sizing software 2100 Expert software | Agilent | Supplies equipment of the 2100 Bioanayzer instrument | |
| Agilent 2100 Bioanalyzer instrument: Test chips | Agilent | Supplies equipment of the 2100 Bioanayzer instrument | |
| Agilent DNA 7500 kit | Agilent | 5067-1506 | For Fragment sizing |
| Agilent DNA 7500 kit: DNA 7500 Ladder (yellow cap) | Agilent | In kit: Agilent DNA 7500 kit (catalog number: 5067-1506) | |
| Agilent DNA 7500 kit: DNA 7500 Markers (green cap) | Agilent | In kit: Agilent DNA 7500 kit (catalog number: 5067-1506) | |
| Agilent DNA 7500 kit: DNA chips | Agilent | In kit: Agilent DNA 7500 kit (catalog number: 5067-1506) | |
| Agilent DNA 7500 kit: DNA Dye Concentrate (blue cap) | Agilent | In kit: Agilent DNA 7500 kit (catalog number: 5067-1506) | |
| Agilent DNA 7500 kit: DNA Gel Matrix Vial (red cap) | Agilent | In kit: Agilent DNA 7500 kit (catalog number: 5067-1506) | |
| Agilent DNA 7500 kit: Electrode Cleaner | Agilent | In kit: Agilent DNA 7500 kit (catalog number: 5067-1506) | |
| Agilent DNA 7500 kit: Spin Filter | Agilent | Supplies of Agilent DNA 7500 kit (catalog number: 5067-1506) | |
| Agilent DNA 7500 kit: Syringe | Agilent | Supplies of Agilent DNA 7500 kit (catalog number: 5067-1506) | |
| Chip priming station | Agilent | 5065-4401 | Supplies equipment of the 2100 Bioanayzer instrument |
| Cubee Mini-centrifuge | GeneReach | aqbd-i | |
| Filter plate vacuum Manifold: MultiScreenHTS Vacuum Manifold | Merck Millipore | MSVMHTS00 | Vacuum instrument for Filter plate vacuum Manifold for PCR product purification |
| Filter plate vacuum Manifold: Silicone stopper | Merck Millipore | XX2004718 | Filter plate vacuum Manifold |
| Filter plate vacuum Manifold: Vacuum pump | Merck Millipore | WP6122050 | Filter plate vacuum Manifold |
| Filter plate vacuum Manifold: Waste collection vessel | Merck Millipore | XX1004705 | Filter plate vacuum Manifold |
| FragilEase Fragile X PCR kit | PerkinElmer | 3101-0010 | For PCR amplification |
| FragilEase Fragile X PCR kit: Sample Diluent | PerkinElmer | In kit: FragilEase Fragile X PCR kit (catalog number: 3101-0010 ) | |
| FragilEase PCR Buffer mix | PerkinElmer | In kit: FragilEase Fragile X PCR kit (catalog number: 3101-0010 ), containing primers. Primer sequences: TCAGGCGCTCAGCTCCGTTTCGGTTTCA (forward) FAM-AAGCGCCATTGGAGCCCCGCACTTCC (reverse) |
|
| FragilEase Polymerase | PerkinElmer | In kit: FragilEase Fragile X PCR kit (catalog number: 3101-0010 ) | |
| FraXsoft analysis software | PerkinElmer | ||
| NanoDrop ND-2000 Spectrophotometer | Thermo Fisher | ||
| Paper towels | |||
| PCR clean up plate: NucleoFast 96 PCR plate | MACHEREY-NAGEL | 743100 | |
| reference DNA sample | Coriell | NA20240 & NA20239 | |
| S1000 96-well Thermal Cycler | Bio-Rad | 1852196 | This can be replaced by other Thermal Cyclers (eg. Veriti™ 96-Well Thermal Cycler, Applied Biosystems, catalog number: 4375786) |
| TriNEST Incubator/Shaker instrument | PerkinElmer | 1296-0050 | |
| UltraPure DNase/RNase-Free Distilled Water | Life Technologies | 10977015 | For 2100 Bioanalyzer electrode cleaning |
| Vortex-Genie 2 | Scientific Industries | SI-0256 (Model G560E) | Conventional vortex mixer |
Referências
- Verkerk, A. J., et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 65, 905-914 (1991).
- Fu, Y. H., et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 67, 1047-1058 (1991).
- Antar, L. N., Li, C., Zhang, H., Carroll, R. C., Bassell, G. J. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Molecular and Cellular Neurosciences. 32, 37-48 (2006).
- Didiot, M. C., et al. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Research. 36, 4902-4912 (2008).
- Bechara, E. G., et al. A novel function for fragile X mental retardation protein in translational activation. PLoS Biology. 7, e16 (2009).
- Ascano, M., et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 492, 382-386 (2012).
- Kenny, P. J., et al. MOV10 and FMRP regulate AGO2 association with microRNA recognition elements. Cell Reports. 9, 1729-1741 (2014).
- Oberle, I., et al. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 252, 1097-1102 (1991).
- Hagerman, R., Lauterborn, J., Au, J., Berry-Kravis, E. Fragile X syndrome and targeted treatment trials. Results and Problems in Cell Differentiation. 54, 297-335 (2012).
- Hatton, D. D., et al. Autistic behavior in children with fragile X syndrome: prevalence, stability, and the impact of FMRP. American Journal of Medical Genetics Part A. 140A, 1804-1813 (2006).
- Mattei, J. F., Mattei, M. G., Aumeras, C., Auger, M., Giraud, F. X-linked mental retardation with the fragile X. A study of 15 families. Human Genetics. 59, 281-289 (1981).
- Backes, M., et al. Cognitive and behavioral profile of fragile X boys: correlations to molecular data. American Journal of Medical Genetics. 95, 150-156 (2000).
- Nolin, S. L., et al. Fragile X analysis of 1112 prenatal samples from 1991 to 2010. Prenatal Diagnosis. 31, 925-931 (2011).
- Nolin, S. L., et al. Expansion of the fragile X CGG repeat in females with premutation or intermediate alleles. American Journal of Human Genetics. 72, 454-464 (2003).
- Fernandez-Carvajal, I., et al. Expansion of an FMR1 grey-zone allele to a full mutation in two generations. Journal of Molecular Diagnostics. 11, 306-310 (2009).
- Terracciano, A., et al. Expansion to full mutation of a FMR1 intermediate allele over two generations. European Journal of Human Genetics. 12, 333-336 (2004).
- Garcia-Arocena, D., Hagerman, P. J. Advances in understanding the molecular basis of FXTAS. Human Molecular Genetics. 19, R83-R89 (2010).
- Juncos, J. L., et al. New clinical findings in the fragile X-associated tremor ataxia syndrome (FXTAS). Neurogenetics. 12, 123-135 (2011).
- Hagerman, R. J., et al. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 57, 127-130 (2001).
- Conway, G. S. Premature ovarian failure and FMR1 gene mutations: an update. Annales d’endocrinologie. 71, 215-217 (2010).
- Conway, G. S., Hettiarachchi, S., Murray, A., Jacobs, P. A. Fragile X premutations in familial premature ovarian failure. Lancet. 346, 309-310 (1995).
- Van Esch, H., Buekenhout, L., Race, V., Matthijs, G. Very early premature ovarian failure in two sisters compound heterozygous for the FMR1 premutation. European Journal of Medical Genetics. 52, 37-40 (2009).
- Bourgeois, J. A., et al. A review of fragile X premutation disorders: expanding the psychiatric perspective. Journal of Clinical Psychiatry. 70, 852-862 (2009).
- Farzin, F., et al. Autism spectrum disorders and attention-deficit/hyperactivity disorder in boys with the fragile X premutation. Journal of Developmental and Behavioral Pediatrics. 27, S137-S144 (2006).
- Hantash, F. M., et al. FMR1 premutation carrier frequency in patients undergoing routine population-based carrier screening: insights into the prevalence of fragile X syndrome, fragile X-associated tremor/ataxia syndrome, and fragile X-associated primary ovarian insufficiency in the United States. Genetics in Medicine. 13, 39-45 (2011).
- Kraan, C. M., et al. FMR1 allele size distribution in 35,000 males and females: a comparison of developmental delay and general population cohorts. Genetics in Medicine. 20 (12), 1627-1634 (2018).
- Saul, R. A., Tarleton, J. C. FMR1-Related Disorders. GeneReviews. , (2012).
- Amos Wilson, J., et al. Consensus characterization of 16 FMR1 reference materials: a consortium study. Journal of Molecular Diagnostics. 10, 2-12 (2008).
- Kwok, Y. K., et al. Validation of a robust PCR-based assay for quantifying fragile X CGG repeats. Clinica Chimica Acta. 456, 137-143 (2016).
- Rousseau, F., Rouillard, P., Morel, M. L., Khandjian, E. W., Morgan, K. Prevalence of carriers of premutation-size alleles of the FMRI gene–and implications for the population genetics of the fragile X syndrome. American Journal of Human Genetics. 57, 1006-1018 (1995).
- Tassone, F., et al. FMR1 CGG allele size and prevalence ascertained through newborn screening in the United States. Genome Medicine. 4, 100 (2012).
- Dombrowski, C., et al. Premutation and intermediate-size FMR1 alleles in 10572 males from the general population: loss of an AGG interruption is a late event in the generation of fragile X syndrome alleles. Human Molecular Genetics. 11, 371-378 (2002).
- Cronister, A., Teicher, J., Rohlfs, E. M., Donnenfeld, A., Hallam, S. Prevalence and instability of fragile X alleles: implications for offering fragile X prenatal diagnosis. Obstetrics and Gynecology. 111, 596-601 (2008).
- Hayward, B. E., Kumari, D., Usdin, K. Recent advances in assays for the fragile X-related disorders. Human Genetics. 136, 1313-1327 (2017).
- Seltzer, M. M., et al. Prevalence of CGG expansions of the FMR1 gene in a US population-based sample. American Journal of Medical Genetics Part B Neuropsychiatrics Genetics. 259B, 589-597 (2012).
- Hayward, B. E., Usdin, K. Assays for determining repeat number, methylation status, and AGG interruptions in the Fragile X-related disorders. Methods in Molecular Biology. , 49-59 (1942).
- Orrico, A., et al. Mosaicism for full mutation and normal-sized allele of the FMR1 gene: a new case. American Journal of Medical Genetics. 78, 341-344 (1998).
- Schmucker, B., Seidel, J. Mosaicism for a full mutation and a normal size allele in two fragile X males. American Journal of Medical Genetics. 84, 221-225 (1999).

